Introduction
Have you heard that the coronavirus “mutates”? Or that there are “several strains” of it around the world? Sounds scary, right? However, the reality is that everything “mutates”. All organisms, over time, acquire differences in their genes, from bacteria to humans. You might be aware that this can happen when your DNA (Deoxyribonucleic Acid) is exposed to UV light (like from the sun!), but this can also happen during DNA replication. This is when a cell uses the template of one of the two DNA strands to make a new complimentary copy of the other strand. Mutation is common to all living organisms (and viruses) and a driver of evolution. This is the first post in a series that will explore coronavirus replication with a focus on the proteins involved.
How does the coronavirus make more of itself?
SARS-CoV-2 uses single-strand Ribonucleic acid (RNA) to encode its genome, not DNA, and hence belongs to a class of “single-strand RNA viruses”. For this reason, the virus needs a different way to copy its genome than “normal” cells have. The viral protein that copies the RNA is called an “RNA-dependent RNA polymerase” (RdRp). This protein uses the viral RNA as a template to make a new copy of viral RNA, by stringing single ribonucleotides together like beads on a string. This process is called polymerization.
A study by the Morse lab at Texas A&M University showed that SARS-CoV-2 RNA polymerase has a remarkable similarity to the RNA polymerase of SARS-CoV (>95%) as well as MERS-CoV [1], the virus which causes Middle-Eastern Respiratory Syndrome. This means that research performed in response to the SARS and MERS epidemics can inform our response to SARS-CoV-2. Unfortunately, a lack of consistent pandemic-preparedness funding means that we didn’t learn as much about RdRp in time as we could have. Still, RNA polymerase might be a viable drug target for halting the spread and reducing the fatality rate of COVID-19.
Structure of the RNA-Dependent RNA Polymerase
By determining the structure of RdRp, and deeply understanding how it works, we can optimize a drug to specifically target it and hinder its function. To this end, in the last few months, several structures of SARS-CoV-2 RNA polymerase have been published.
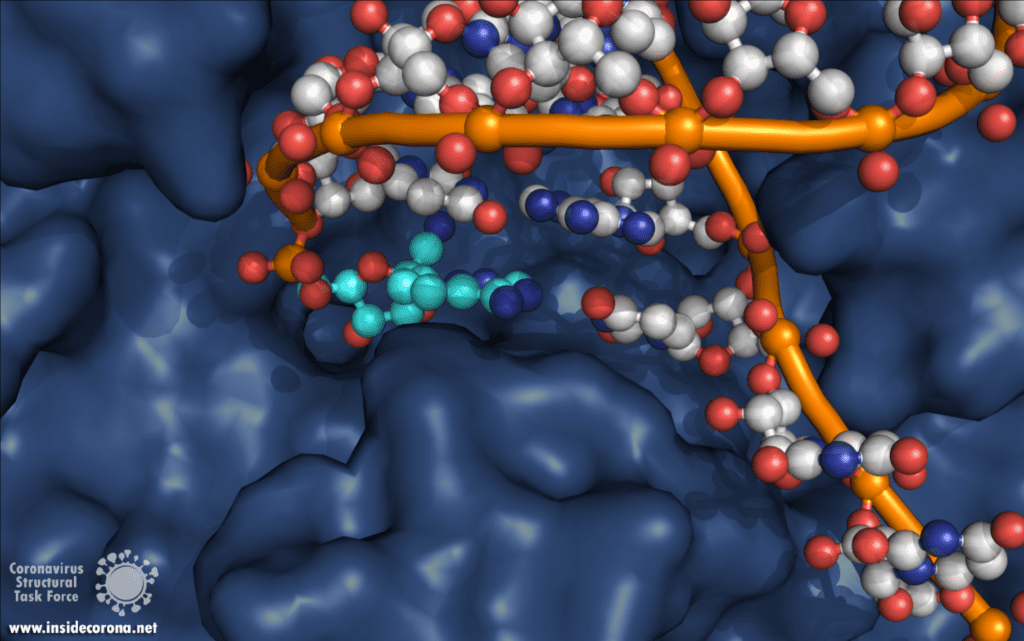
One interesting structure shows RNA polymerase in action, in the process of elongating an RNA strand (see Figure 1).[2] This structure clearly show the polymerase in complex with smaller proteins, non-structural protein 7 and 8 (nsp7 and nsp8). These proteins improve how well the RNA polymerase binds the template RNA and also how long it stays bound before dissociating – a feature called “processivity”.[3]
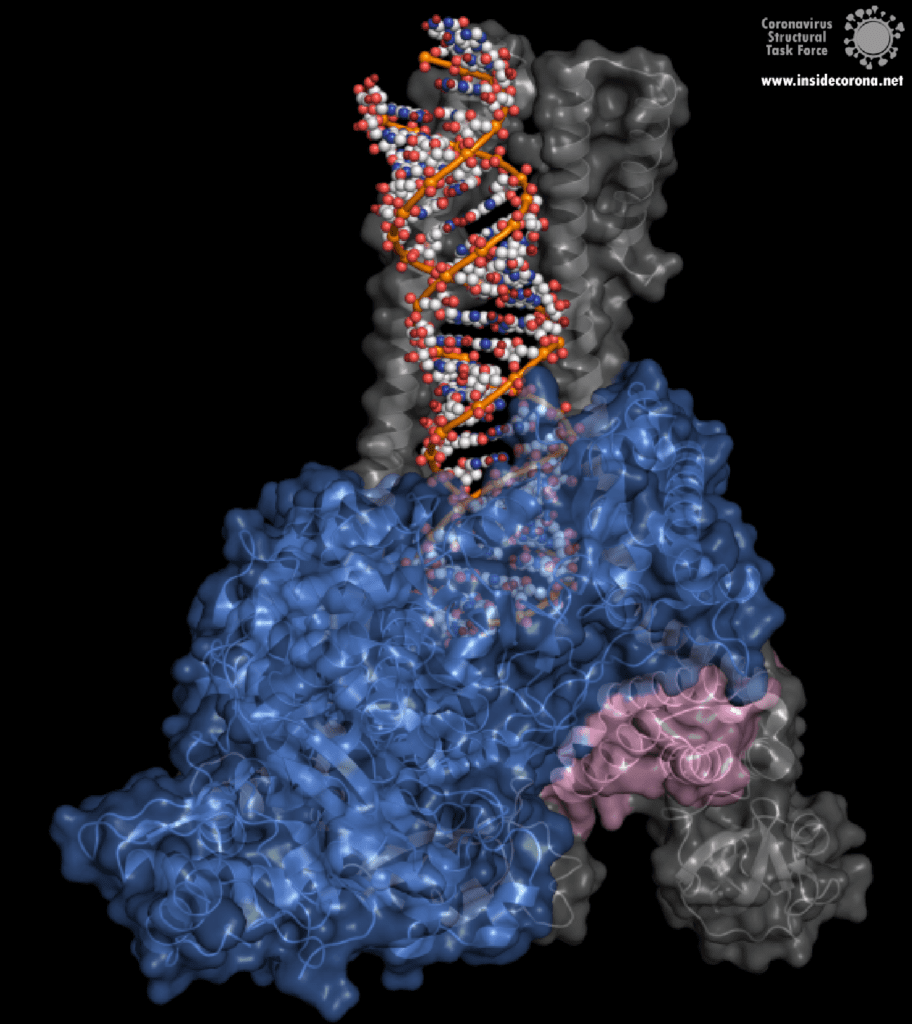
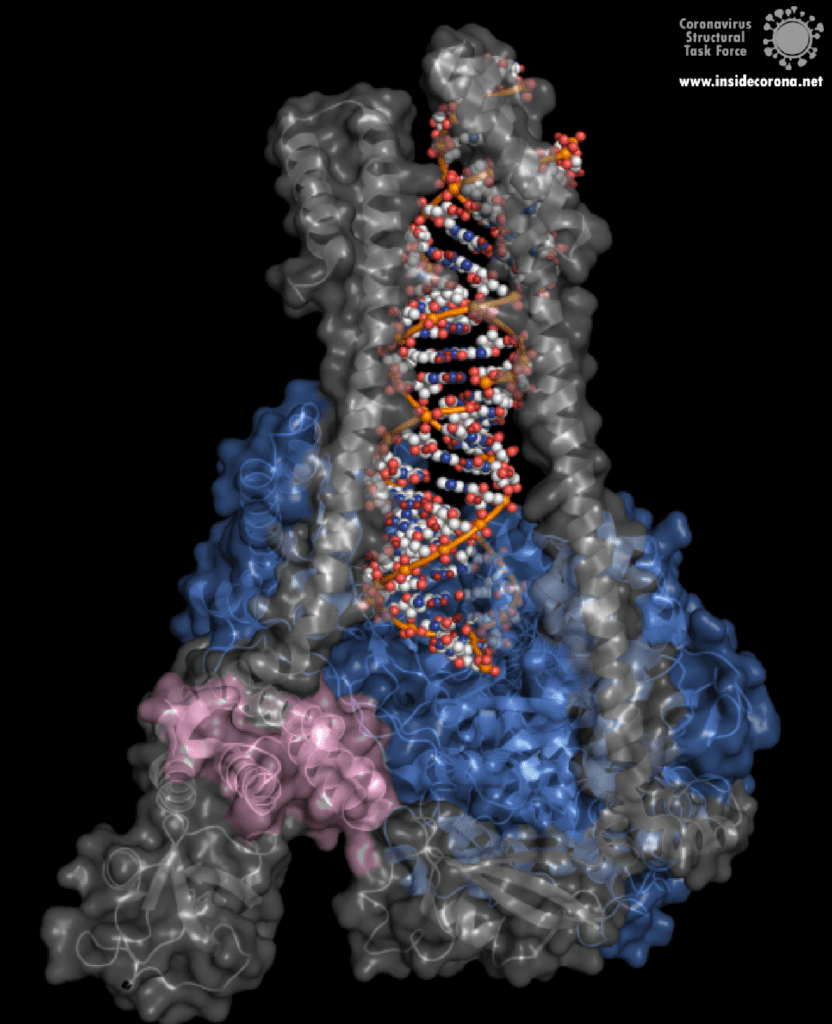
In the center of the protein is the area where the main action happens, called the “active site”. The amino acids of the polymerase that form the active site have a particular shape and chemical properties, which enable the polymerization reaction to occur very rapidly. In fact, the polymerase can string together as many as 100 nucleotides per second! [3] New RNA molecules can enter the active site through a little window to be added to the growing RNA chain. It is here that the antiviral drugs make their move!
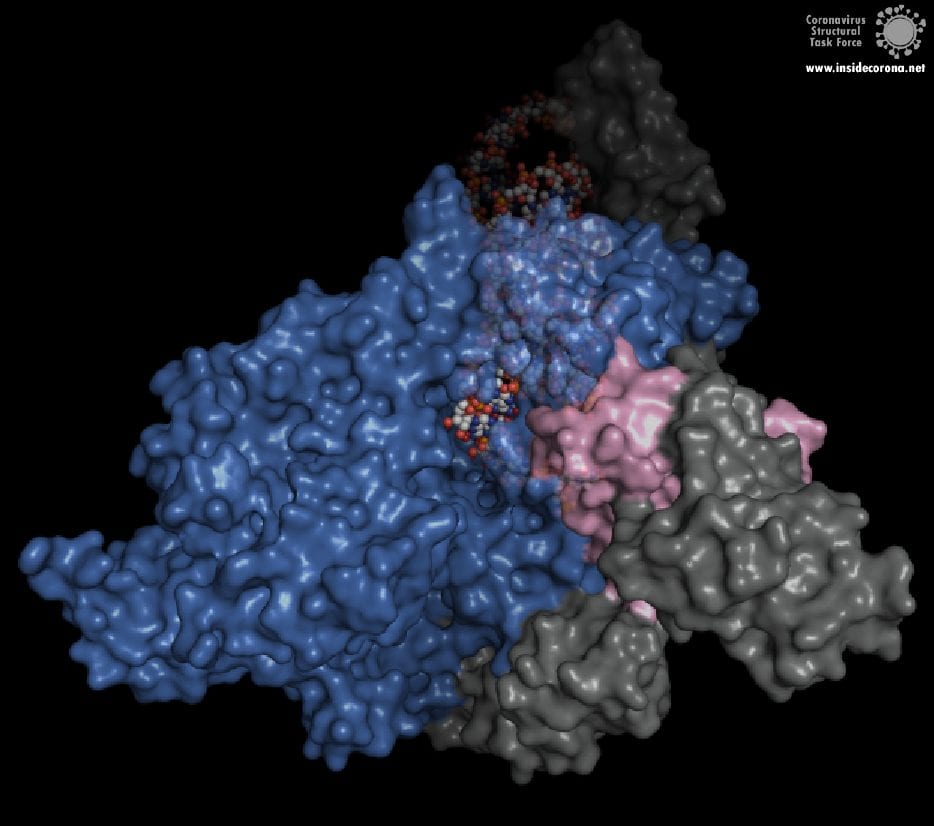
How do antiviral drugs attack RNA-dependent RNA polymerase?
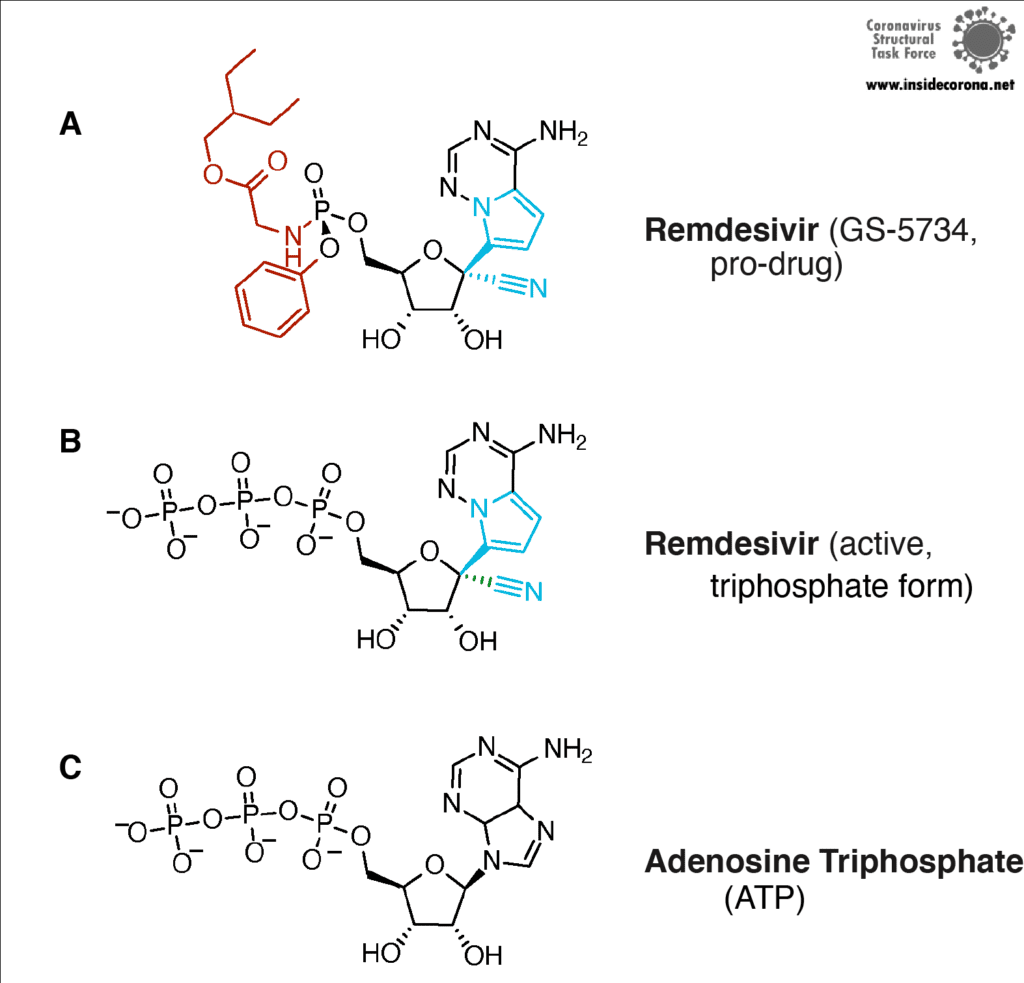
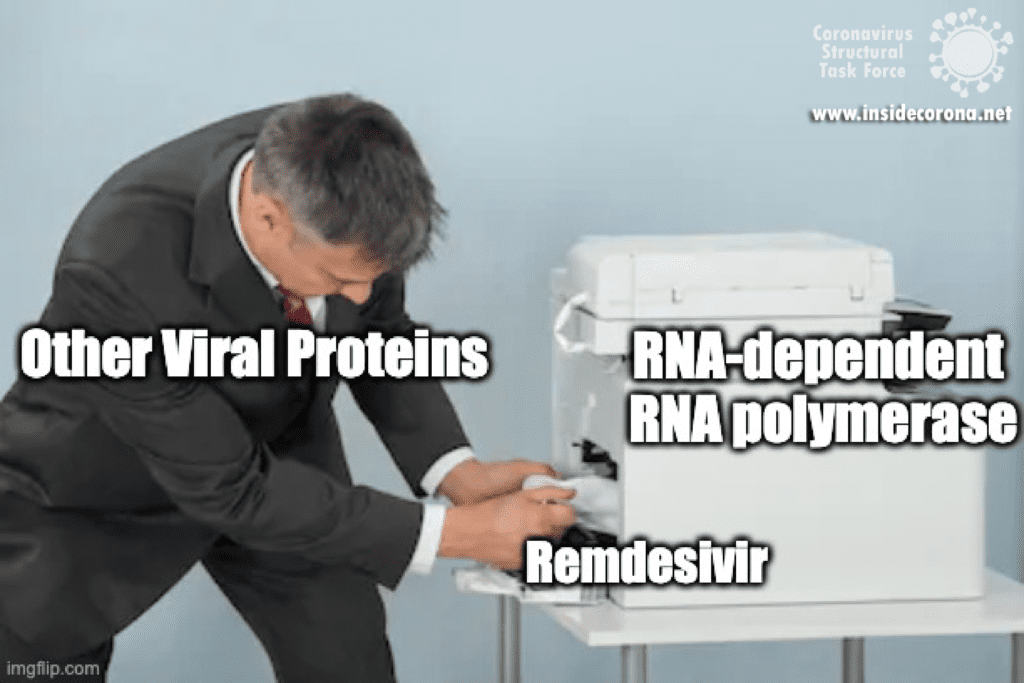
First, let’s talk about Gilead’s FDA-approved drug, Remdesivir, which has taken the spotlight in the search for COVID-19 cures. Remdesivir (which has a fancy chemistry ID, GS-5734, and is sold under the brand name Veklury), is a “nucleotide analog”, which means that it mimics the shape and chemistry of the nucleotides that make up RNA and DNA (see figure).
Remdesivir was developed originally as a general antiviral drug and was later shown to protect cells (in a test tube) and monkeys (not in a test tube) from the Ebola Virus [4]. However, this was recent enough, and science is slow enough that, until the COVID-19 pandemic, large-scale clinical trials of Remdesivir hadn’t been done yet. So scientists and doctors have been rushing to test the drug in COVID-19 patients. In fact, the US and Japan both approved the drug for “Emergency Use Authorization'' for severe COVID-19 patients as early as May [5], [6]. And, in July, the European Medicines Agency gave Remdesivir a “conditional marketing authorization” (used for drugs that meet an unmet medical need but have insufficient data for normal approval). This allows the use of Remdesivir in severe COVID-19 patients through the next year [7]. So, how the heck does a drug for Ebola, Influenza, or some other viruses also work against COVID-19? I was concerned by this when the news about all the drug trials were coming out – and I’m sure I wasn’t the only one...
The simple answer to that is all these viruses need to do the same thing - copy their RNA genome from an RNA template. And in order to do that, they all end up using basically the same tool, an RNA-Dependent RNA polymerase. And all drugs that are nucleotide analogs use the very same trick: they dress up like ribonucleotides (the "beads on a string" from before) and fool the RNA polymerase into letting them into the active site. Once inside, they get “stuck” in the active site, jamming the polymerase machine. Since this trick should work for any viral RNA polymerase, we can use these drugs for any RNA virus, and call them ‘general antivirals’. Of course, in practice, this doesn't always work, because there are differences between the different RNA polymerases. However, it is a great place to start! In the future, if we have general antivirals for SARS-CoV-2 all ready-to-go, we may be better equipped to deal with another coronavirus outbreak!

The Chemistry of Remdesivir
Remdesivir resembles the nucleotide adenine in structure, although it has some fancy chemical add-ons which help make it a better drug (thank you, medicinal chemistry!). When Remdesivir is injected into a vein, it travels through the bloodstream and enters into our cells, which recognize it as a foreign substance and try to digest it. However, what ends up happening is that the cells remove just the fancy chemical add-ons, and then confuse it for a normal adenine nucleotide. In infected cells, the viral RNA-dependent RNA polymerase then starts grabbing these molecules and inserting them into the new viral RNA strand in place of adenine molecules. Remdesivir, now attached to the RNA, jams the polymerase, rendering the virus unable to make more copies of its genome. Ultimately, this halts viral replication and helps the patient fight off the virus.
Another drug that inhibits the RNA polymerase activity is Favipiravir, sold under brand names Avigan, Abigan, and FabiFlu. Favipiravir has been discovered by Toyama Chemical Co., Ltd. in Japan and it has a similar mechanism to Remdesivir, except that it mimics a guanosine nucleoside instead of an adenine nucleotide [8]. This drug was approved in Japan back in 2014 for use in resistant cases of Influenza A and B, but still remains unapproved in the US (still in Phase II and Phase III clinical trials) and the UK [9]. This drug is also being tested for use against Ebola virus, Lassa virus, and currently SARS-CoV-2 in 43 countries. The approval of Favipiravir for COVID-19 has been much faster in China (Mar 15, 2020), Russia (Jun 3, 2020), and India (Jun 20, 2020)[10], [11]. Nonetheless, other countries, including Japan, are in various stages of clinical trials, and the results are anticipated to be out by the end of July [10].
So...do we have a cure for SARS-CoV-2?
Sadly, not yet. While the speed at which Remdesivir has gone through clinical trials is unprecedented, more work needs to be done to make sure it is safe and effective. Since (in the big scope of things) not a lot have people have taken Remdesivir, we aren’t really sure what all the side effects are, although there is emerging evidence for liver and kidney damage [12, 13]. The most common side effects are nausea (10% and 9% of patients), indigestion (7%) and increase of transaminases (6% and 8%). In one study, 3.6% of patients in a 10-day trial needed to stop taking therapy due to the latter. However, serious viral infections can also cause liver damage, so separating the two causes is a challenge! Remdesivir is not a cure-all, either. In one study it improved the recovery time from 15 days to 11 days, but it showed no effect for patients with mild to moderate disease, and no difference in median recovery time for patients who were already on a ventilator [14]. Since the drug has to be given by infusion over several days, there is a pretty small window in which Remdesivir can actually help.
Likewise, Favipiravir has its own side effects such as liver damage, elevated uric acid levels, kidney damage, skin allergies, etc. [15]. These effects restrict it for use by severe diabetes and heart patients. On top of that, it is not suitable for pregnant women because it can cause potential fetal deaths and deformities. It has been shown that Favipiravir works only during the earlier stages of SARS-CoV-2 infection when the body’s immune system isn’t totally drained, whereas it can result in a cytokine storm (when your immune system really freaks out) in severely ill patients. But, unfortunately, the virus doesn’t differentiate between humans while attacking, so a universal drug for COVID-19 has to be safe for use by all people.
However, these drugs are better than nothing, and by understanding the mechanisms involved, scientists can continue to improve upon the existing drugs for the benefit of all. While most of the ‘general antivirals’ that target RNA Polymerase have failed with SARS-CoV-2, Remdesivir has been relatively successful. Scientists think that this is actually because of a proofreading protein in SARS-CoV-2 called exonuclease. Immediately after the RNA-polymerase makes new RNA, exnuclease checks to make sure the new RNA is correct. In one study, another drug that mimics RNA called Ribivarin was shown to be removed from newly synthesized RNA by exonuclease [16]. Thankfully, Remdesivir is not excised , which is likely why it has been more successful than the other options [17], [18]. To read more about how nsp14 maintains the integrity and virulence of SARS-CoV-2, tune in to a future blog entry!
Recommended Structures
For those interested in reviewing the structures further, they are available in our GitHub repo, along with information about validation and, where relevant, improved structures. For a high-resolution comparison of the active site with and without Remdesivir, 7BV2 and 7BV1 (respectively) were published together at 2.5 and 2.8 Å. The elongating structure of the complex shown above (6YYT) has the polymerase as well as the cofactors and RNA very well resolved, with little "missing" density and a resolution of 2.9 Å. It is likely preferable to 6M71 and 7BTF, which were published with a similar resolution but with less of the complex resolved, and no RNA. For those interested, 7C2K and 7BZF (at 2.93 Å and 3.26 Å) show the complex bound to RNA in a pre- and post-translocation state.
Sources
[1] J. S. Morse, T. Lalonde, S. Xu, and W. R. Liu, “Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV,” ChemBioChem, vol. 21, no. 5, pp. 730–738, Mar. 2020, doi: 10.1002/cbic.202000047.
[2] H. S. Hillen, G. Kokic, L. Farnung, C. Dienemann, D. Tegunov, and P. Cramer, “Structure of replicating SARS-CoV-2 polymerase,” Nature, May 2020, doi: 10.1038/s41586-020-2368-8.
[3] W. Yin et al., “Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir,” Science, p. eabc1560, May 2020, doi: 10.1126/science.abc1560.
[4] R. T. Eastman et al., “Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19,” ACS Cent. Sci., May 2020, doi: 10.1021/acscentsci.0c00489.
[5] O. of the Commissioner, “Coronavirus (COVID-19) Update: FDA Issues Emergency Use Authorization for Potential COVID-19 Treatment,” FDA, May 04, 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment (accessed Jul. 08, 2020).
[6] A. Sternlicht, “Japan Approves Remdesivir For Use On Severe COVID-19 Patients,” Forbes. https://www.forbes.com/sites/alexandrasternlicht/2020/05/07/japan-approves-remdesivir-for-use-on-severe-covid-19-patients/ (accessed Jul. 08, 2020).
[7] D. CZARSKA-THORLEY, “First COVID-19 treatment recommended for EU authorisation,” European Medicines Agency, Jun. 25, 2020. https://www.ema.europa.eu/en/news/first-covid-19-treatment-recommended-eu-authorisation (accessed Jul. 10, 2020).
[8] E. De Clercq, “New Nucleoside Analogues for the Treatment of Hemorrhagic Fever Virus Infections,” Chem. Asian J., vol. 14, no. 22, pp. 3962–3968, Nov. 2019, doi: 10.1002/asia.201900841.
[9] K. Shiraki and T. Daikoku, “Favipiravir, an anti-influenza drug against life-threatening RNA virus infections,” Pharmacol. Ther., vol. 209, p. 107512, May 2020, doi: 10.1016/j.pharmthera.2020.107512.
[10] T. Hornyak, “Japan sending Fujifilm’s flu drug favipiravir to over 40 countries for Covid-19 trials,” CNBC, May 04, 2020. https://www.cnbc.com/2020/05/04/fujifilms-flu-drug-favipiravir-sent-to-43-nations-for-covid-19-trials.html (accessed Jul. 14, 2020).
[11] G. P. Ltd, “Glenmark Becomes the First Pharmaceutical Company in India to Receive Regulatory Approval for Oral Antiviral Favipiravir, for the Treatment of Mild to Moderate COVID-19.” https://www.prnewswire.com/in/news-releases/glenmark-becomes-the-first-pharmaceutical-company-in-india-to-receive-regulatory-approval-for-oral-antiviral-favipiravir-for-the-treatment-of-mild-to-moderate-covid-19-855346546.html (accessed Jul. 14, 2020).
[12] Goldman, J. D. et al. Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. N. Engl. J. Med. (2020) doi:10.1056/NEJMoa2015301
[13] Remdesivir Safety Forecast: Watch the Liver, Kidneys | MedPage Today. https://www.medpagetoday.com/infectiousdisease/covid19/86582
[14] J. H. Beigel et al., “Remdesivir for the Treatment of Covid-19 — Preliminary Report,” N. Engl. J. Med., vol. 0, no. 0, p. null, May 2020, doi: 10.1056/NEJMoa2007764.
[15] Sandhya Ramesh, “Favipiravir, Japanese drug that’s the new Covid treatment hope your chemist will soon stock,” ThePrint, Jun. 25, 2020. https://theprint.in/health/favipiravir-japanese-drug-thats-the-new-covid-treatment-hope-your-chemist-will-soon-stock/447987/ (accessed Jul. 14, 2020).
[16] F. Ferron et al., “Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA,” Proc. Natl. Acad. Sci., vol. 115, no. 2, pp. E162–E171, Jan. 2018, doi: 10.1073/pnas.1718806115.
[17] C. J. Gordon, E. P. Tchesnokov, J. Y. Feng, D. P. Porter, and M. Gotte, “The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus,” J. Biol. Chem., Feb. 2020, doi: 10.1074/jbc.AC120.013056.
[18] L. Zhang et al., “Role of 1’-Ribose Cyano Substitution for Remdesivir to Effectively Inhibit both Nucleotide Addition and Proofreading in SARS-CoV-2 Viral RNA Replication,” bioRxiv, p. 2020.04.27.063859, Apr. 2020, doi: 10.1101/2020.04.27.063859.
