Introduction
The novel Coronavirus SARS-CoV-2 incorporates various structural proteins in its protective coat. In order to find a potential drug target against the spreading pandemic, a lot of scientific research focusses on the characteristic spike glycoprotein as a therapeutic target. But apart from the spikes, several other structural proteins were found to decorate the virus hull of which the envelope protein (E protein) is the smallest one, consisting of only 75 amino acids. Even though it is an integral membrane protein, the envelope protein is also localized in the host ER, Golgi, and ERGIC (ER-Golgi intermediate compartment) [1], where it is essential for virus formation.
Interestingly, research on this protein can shed light on the origin of the novel coronavirus, which currently dominates everyday life all over the globe. Sequence comparisons of several different envelope proteins strengthen the assumption that SARS-CoV-2 may originate from Bat-CoV or Pangolin-CoV due to a high sequence homology [2]. The E protein of SARS-CoV-2's "older brother" SARS-CoV exhibits a nearly identical sequence with 91% homology [2] as well and has been structurally determined based on nuclear magnetic resonance (NMR) data. Yet, until now, solving the 3D structure of SARS-CoV-2 E protein turns out to be quite challenging, and hence no experimental structure is available for the new coronavirus [3].
Structure comparison with SARS-CoV E protein
We, as structural biologists aim to uncover and refine the structures of as many of the novel virus's proteins as possible. But, as long as no structures of SARS-CoV-2 E protein have been solved, this could only be achieved by comparing it to the existing structures of SARS-CoV envelope protein.
Topology and structural features
The topology of SARS-CoV E protein is mainly separated into three domains: A short hydrophilic N-terminus, that has an identical sequence in SARS-CoV-2 [3] and works as a Golgi-targeting signal; a long mainly hydrophobic transmembrane domain (TMD), and a long hydrophilic C-terminal domain. Studies on the question whether the C- and the N-terminus are luminal or cytoplasmic have had different results, suggesting that the E protein’s topology could differ depending on its multiple functions [3].
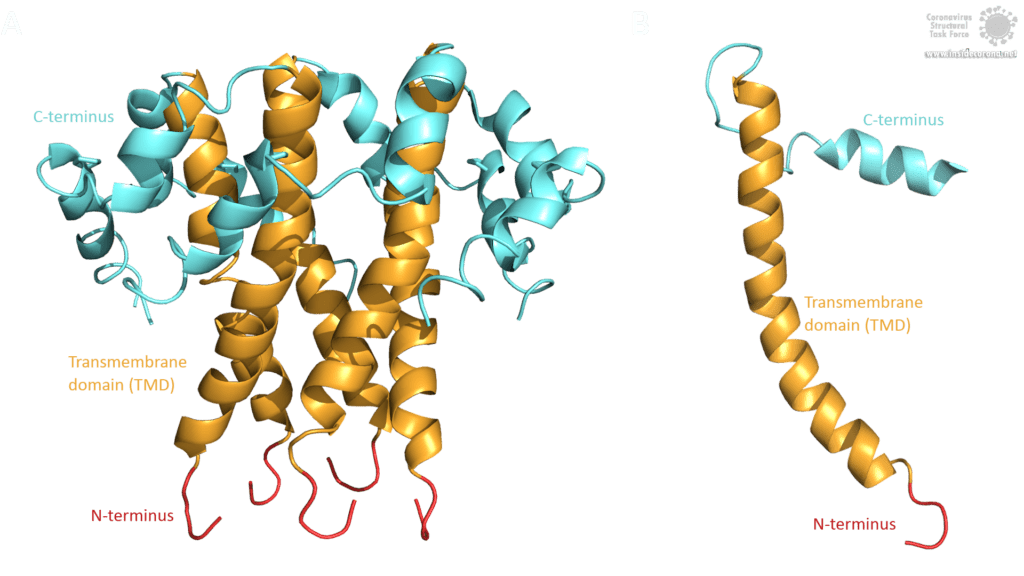
Image by Luise Kandler
The E protein of SARS-CoV comprises several interesting structural features: A long α-helix with amphipathic parts forms the Transmembrane domain (TMD). The C-terminus, however, incorporates a short α-helix which is believed to be in a dynamic equilibrium with a less abundant β-coil-β-motif. Both helices are connected by a turn [4]. The β-coil-β-motif with a conserved proline residue (Pro-54) has been proposed to function as Golgi targeting signal, and to switch its conformation in order to alter the E protein's function in the host cell [4]. Furthermore, the C-terminus contains a PDZ-binding motif (PBM) at residues 73-76 [3]. This PBM domain slightly differs in coronaviruses but a DLLV motif is conserved in the E proteins of SARS-CoV, Bat-CoV, and SARS-CoV-2 [2]. Unfortunately, there are no PDB structures available that exhibit the β-coil-β-motif nor the PBM domain.
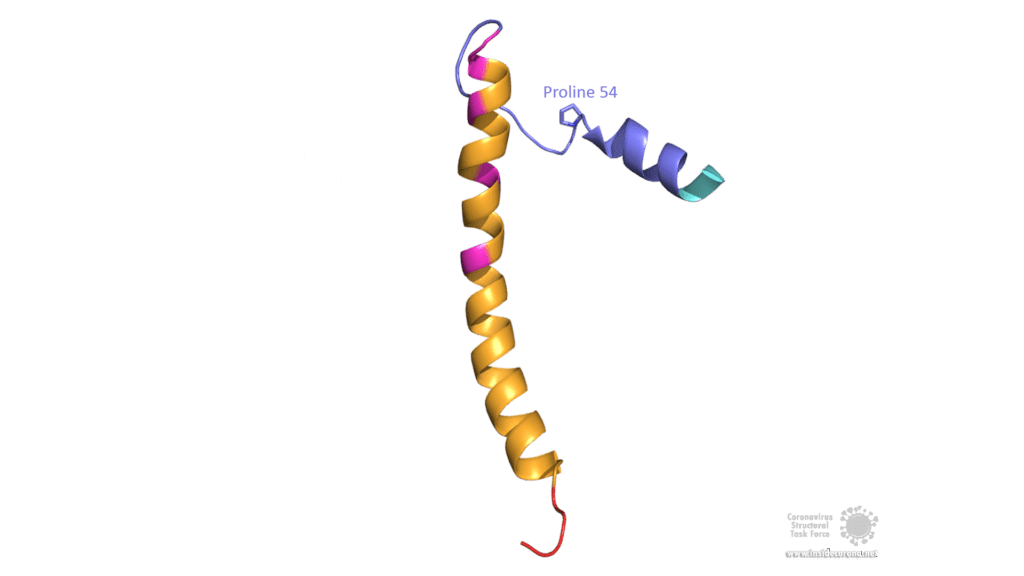
Image by Luise Kandler
Structural variants - Oligomerization and posttranslational modifications
The E protein comes in two different forms. Apart from a monomeric structure, the protein also oligomerizes to form a pentameric viroporin in the host cell's Golgi membrane. Whether the E proteins that are embedded in the viral hull are pentamers or monomers is not yet clear. Oligomerization is induced by the amphipathic α-helix of the TMD [3] and is proposed to be mainly mediated by residue Val-25 as well as residue Asn-15 being slightly involved [3]. Both residues are conserved in SARS-CoV-2 as well. To anchor the pore in the Golgi membrane, the hydrophobic amino acids of the TMD orientate towards the phospholipids. Additionally, basic positively charged residues interfere with the negatively charged phospholipids via electrostatic interactions [3].
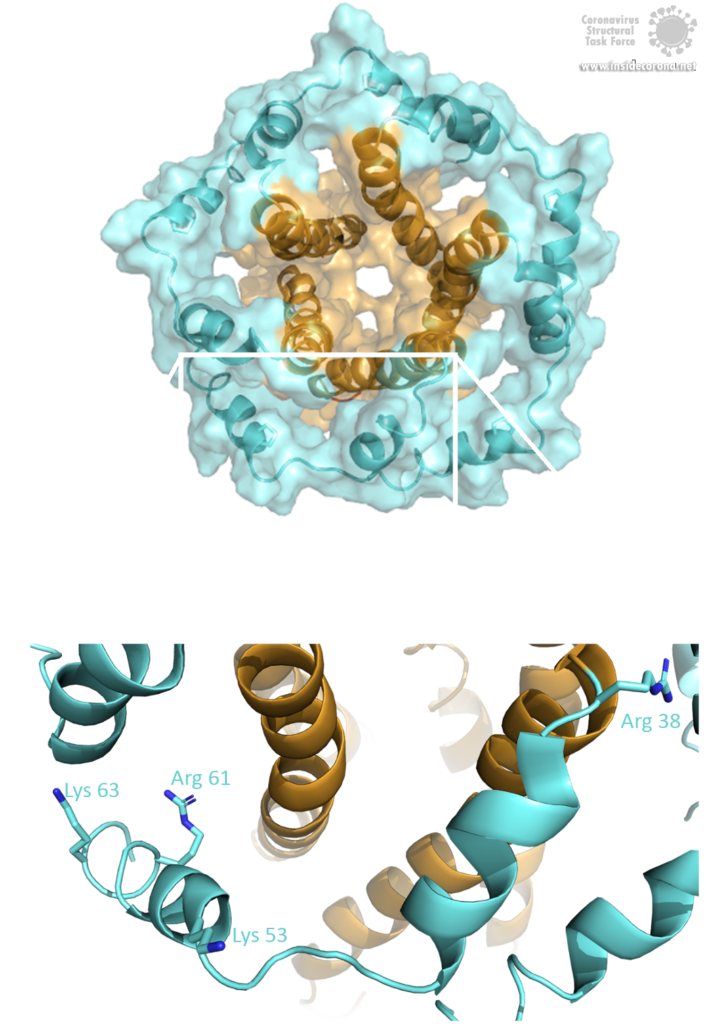
Image by Luise Kandler
Other structural variants are obtained by posttranslational modifications, which have been detected in the E protein of SARS-CoV and other coronaviruses. Palmitoylation is the addition of palmitic fatty acid to cysteine residues which increases the protein's hydrophobicity. Hence, the palmitoylation of E protein assists in membrane anchoring and probably aids Golgi targeting. Ubiquitination of the E protein might function as negative regulation of E protein levels [3]. It has been shown that the optimal amount of E protein present in the host cell is important for a successful production of new viruses. Another modification, namely glycosylation, adds oligosaccharide fragments to asparagine residues in a certain motif (Asn-X-Ser/Thr) which is also conserved in E protein. In SARS-CoV, residue Asn-66 embedded in the motif Asn-Ser-Ser was proven to be glycosylated. This may help to recruit chaperone proteins of the host cell to aid in the correct folding of newly synthesized viral proteins as well as in defense against the host immune system. Experimental data suggest that glycosylation of Asn-66 might also promote E protein's monomeric functions as it prevents oligomerization [3].
Connecting function and structure
To understand a molecule's biological function is the main goal of experimental structure determination. A viral protein can be targeted by drugs best if the atomic structure is known. The envelope protein has various structural conformations and thus multiple functions, both as a monomer and as a pentamer.
Monomeric functions: Golgi-targeting and viral assembly
The E protein comprises a Golgi-targeting signal in the β-coil-β motif of the C-terminus and another one in the N-terminal domain. Additionally, palmitoylation is believed to be involved in this function. Accordingly, after being translated at the ER, the E protein is located to the Golgi membrane. From there, the virus acquires the membrane for a new viral envelope [3]. Once the protein is located to the Golgi, one of its main functions as a monomer is in viral assembly, which means the process of gathering all the viral macromolecules (proteins and the RNA genome) to form a virus-like particle. During this assembly, the virus-like particle buds into the lumen of ERGIC and follows the way through the host cell's secretory pathway. Several experiments confirm the involvement of the envelope protein together with the membrane protein (M) into this process. It has been proposed that the E protein rather induces membrane curvature and scission, whereas the M protein may coordinate viral assembly. Nevertheless, SARS-CoV-infected cells still produce virus-like particles in the absence of E protein, but virus trafficking to the cell surface and viral secretion are hampered, resulting in a lower number of mature virions, an atypic morphology and a higher rate of propagation incompetent virions [3]. Further investigation will be necessary to analyze the exact mechanism behind the membrane formation of virions.
After finding its way through the secretory pathway, the mature virion is released from the host cell. The process of detaching from the host membrane is known as scission and is either coordinated by the virus's own scission proteins or by the host cell's scission machinery (called ESCRT). Which one is the case for SARS-CoV-2 is still unclear. Infected cells lacking the scission machinery exhibit a “beads-on-a-string” morphology, with the virions being stuck to the host membrane in an elongated shape. This morphology was found in influenza-infected cells lacking the M2 protein, which proves that M2 is involved in this scission process. Given that SARS-CoV E protein is suggested to be functionally equivalent to M2, due to similar structural features, the E protein is proposed to be involved in the scission process as well [3].

Image by Ann (Hui) Liu, https://animationlab.utah.edu/ [5]
Pentameric functions: Ion-channel activity
While located at the Golgi, some of the SARS-CoV E proteins oligomerize and form a pentameric viroporin. These pores of SARS-CoV E protein function as ion channels. They mainly favor the transport of Na+ and K+, but were also found to be permeable for Ca2+ ions and eventually for H+ ions. Even though the primary purpose of transporting cations is not yet clear, Ca2+ is proposed to trigger the inflammatory response seen in acute respiratory distress syndrome [7].
Residue Asn-15 has been suggested to act as a "filter" for this ion selectivity [6], which can further be affected by the charge of the membrane's lipid head group. Deletion of the envelope protein in its pentameric form demonstrates that ion channel activity is not essential for viral replication, but yet attenuates the virulence [8].
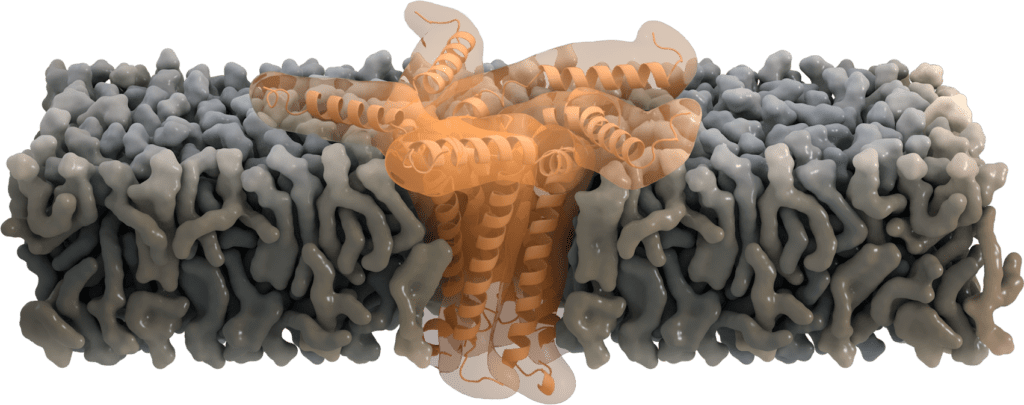
Image by Thomas Splettstoesser, www.scistyle.com
Pathogenesis and the E protein as a potential drug target
Interactions of viral proteins with host cell proteins de-regulate many physiological processes. In patients suffering from SARS-CoV infections, these de-regulating protein-protein interactions greatly contribute to pathogenesis. Some of the observed symptoms are also present in a SARS-CoV-2 infected patient.
Interactions of the envelope protein with proteins of the host cell are mediated by its PDZ-binding motif (PBM) at the very end of the C-terminus. The motif binds to the PDZ domain of adaptor proteins, which are subsequently bound by other cellular proteins, activating a signaling cascade that may result in pathogenesis. Some of these interactions were proposed or even proven to induce symptoms like lymphopenia [9], changes in fluid volume, blood pressure, and water homeostasis, as well as tissue damage, edema and acute respiratory distress syndrome (ARDS) [10], due to an overexpression of inflammatory cytokines (which are also regulated by the leader protein nsp1). Another protein-protein interaction was found to disrupt tight junctions of pulmonary epithelial cells in the lungs. This eventually results in an epithelial barrier failure and virions breaking through the alveolar wall causing a systemic infection [11]. O. Wittekindt writes [12]: "The breakdown of the epithelial barrier is a hallmark in respiratory distress syndromes (...)" Furthermore, the ion channel activity of the E protein activates the inflammatory pathway by channeling Ca2+ resulting in lung damage in infected mice [7]. Inhibition of the viroporin by hexamethylene amiloride (HMA) [8] reduces the activation of the inflammasome, which makes the ion channel of E protein a potential therapeutic target. Additionally, as a part of the host cell's viral defense, the ER stress response is activated, once the protein folding capacity of the ER is overloaded by additional expression of viral proteins. This can lead to apoptosis of the host cell. However, experiments confirm that the E protein contributes to pathogenesis by suppressing the ER stress response to maintain the survival of the host cell [3].
As a potential target for drug treatment, protein-protein interactions of the E protein are quite promising. Its PBM domain can bind cellular proteins that are involved in pathogenesis. Experimental truncation of this domain shows that it may be possible to find a live vaccine with a mutated but intact PBM and thus attenuated pathogenicity. Identifying more interacting partners could provide a more targeted therapy, though. The absence of E protein furthermore leads to reduced viral titers, crippled viral maturation, and propagation-defective progeny [3], making E protein-deficient virions also a potential vaccine candidate.
In conclusion, one could say that the E protein of SARS-CoV-2 is another valuable drug target. While the protein's "older brother" SARS-CoV E protein gives us much insight into its function, an experimental structure determination of SARS-CoV-2 E protein would be highly desirable. Until then, the envelope protein SARS-CoV-2 remains a small but mysterious structure.
Best PDB structures available
- 2MM4: This NMR structure is a monomer of SARS-CoV envelope protein (E). It covers the transmembrane domain completely and the C- and N-terminus partly. The structure is involved in membrane curvature, membrane scission, and viral assembly.
- 5X29: This NMR structure is an oligomer of SARS-CoV envelope protein (E). The structure is a pentamer of five identical monomers. It covers the transmembrane domain completely and the C- and N-terminus partly. This structure functions as a membrane-anchored ion channel.
Literature
[1] J. Nieto-Torres, M. DeDiego, E. Álvarez, J. Jiménez-Guardeño, J. Regla-Nava, M. Llorente, et al.: Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein, Virology, 2011
[2] M. Bianchi, D. Benvenuto, M. Giovanetti, S. Angeletti, M. Ciccozzi, S. Pascarella: Sars-CoV-2 Envelope and Membrane proteins: differences from closely related proteins linked to cross-species transmission, Preprint, 2020
[3] D. Schoeman, B. Fielding: Coronavirus envelope protein: current knowledge, Virology Journal, 2019
[4] Y. Li, W. Surya, S. Claudine, J. Torres: Structure of a Conserved Golgi Complex-targeting Signal in Coronavirus Envelope Proteins, The Journal Of Biological Chemistry, 2014
[5] Ann (Hui) Liu, in https://animationlab.utah.edu/
[6] K. Pervushin, E. Tan, K. Parthasarathy, X. Lin, F. Jiang, D. Yu, A. Vararattanavech, T. Soong, D. Liu, J. Torres: Structure and Inhibition of the SARS Coronavirus Envelope Protein Ion Channel, PloS Pathogens, 2009
[7] J. Nieto-Torres, C. Verdiá-Báguena, J. Jimenez-Guardeño, J. Regla-Nava, C. Castaño-Rodriguez, R. Fernandez-Delgado, et al.: Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome, Virology, 2015
[8] J. Nieto-Torres, M. DeDiego, C. Verdiá-Báguena, J. Jimenez-Guardeño, J. Regla-Nava, R. Fernandez-Delgado, et al.: Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis, PLoS Pathogens, 2014
[9] Y. Yang, Z. Xiong, S. Zhang, Y. Yan, J. Nguyen, B. Ng, et al.: Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors, Biochemical Journal, 2005
[10] J. Jimenez-Guardeño, J. Nieto-Torres, M. DeDiego, J. Regla-Nava, R. Fernandez-Delgado, C. Castaño-Rodriguez, et al.: The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis, PLoS Pathogens, 2014
[11] K. Teoh, Y. Siu, W. Chan, M. Schlüter, C. Liu, J. Peiris, et al.: The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis, Mol Biol Cell, 2010
[12] O. Wittekindt: Tight junctions in pulmonary epithelia during lung inflammation, Springer Verlag, 2016
