Für diesen Beitrag exisitiert leider keine deutsche Übersetzung.
This article has been written for the journal Crystallographic Reviews and has been published on 11 June 2022 online. For additional information, data and tables, please see the full publication: https://doi.org/10.1080/0889311X.2022.2065270.
Abstract
The SARS-CoV-2’s endoribonuclease (NendoU) nsp15, is an Mn2+ dependent endoribonuclease specific to uridylate that SARS-CoV-2 uses to avoid the innate immune response by managing the stray RNA generated during replication. As of the writing of this review 20 structures of SARS-CoV-2 nsp15 have been deposited into the PDB, largely solved using X-ray crystallography and some through Cryo-EM. These structures show that a nsp15 monomer consist of three conserved domains, the N-terminal oligomerization domain, the middle domain, and the catalytic NendoU domain. Enzymatically active nsp15 forms a hexamer through a dimer of trimers (point group 32), whose assembly is facilitated by the oligomerization domain. This review summarises the structural and functional information gained from SARS-CoV-2, SARs-CoV and MERS-CoV nsp15 structures, compiles the current structure-based drug design efforts, and complementary knowledge with a view to provide a clear starting point for downstream structure users interested in studying nsp15 as a novel drug target to treat COVID-19.
Introduction
SARS-CoV-2 is a nidovirus with a non-segmented positive-sense RNA genome, meaning the RNA genome is read from 5′ to 3′ and can be directly translated into viral proteins; it’s effectively messenger RNA. The RNA genome of SARS-CoV-2 is one of the largest RNA genomes among RNA viruses [1], comprised of a replicase gene which encodes non-structural proteins (nsps), structural proteins, and accessory proteins. The genome can produce two different polyprotein chains through a ribosomal frameshift [2] (ORF1a and ORF1b). Once translated, these polyproteins are cleaved by one of the two encoded proteases (3C-like protease (nsp5) or papain-like-protease (nsp3)) to yield between 15 and 16 non-structural proteins, which assemble into a large membrane-bound replicase complex (RTC).
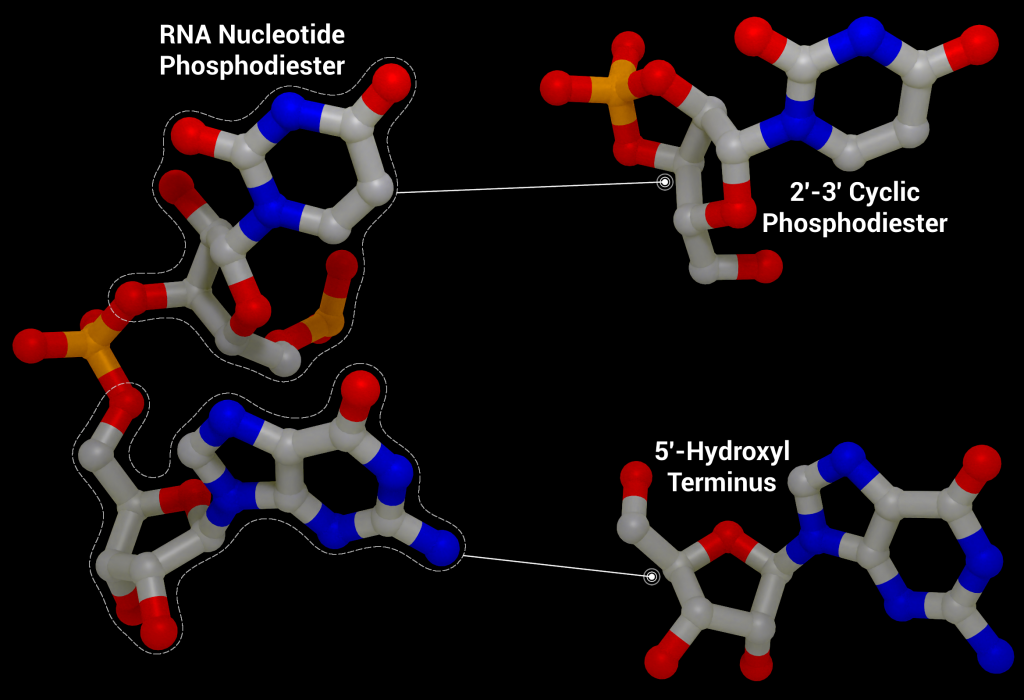
One of these non-structural proteins is nsp15, a 346 amino acid nidoviral RNA uridylate‐specific and Mn2+-dependent [3] endoribonuclease (NendoU). Its gene is found towards the end of the non-structural proteins in the SARS-CoV-2 genome on ORF1b (bases 6453 to 6798[4]). Nsp15 preferentially cleaves the 3′ end of uridine, producing a 2′‐3′ cyclic phosphodiester and 5′‐hydroxyl terminus [5] (Figure 1). Nsp15 is conserved across coronavirus family members [6], to the point where it has been proposed as a universal genetic marker to distinguish nidoviruses [7] from all other RNA virus families. Although highly conserved (88% sequence identity with SARS-CoV-2, 50% with MERS, and 43% with HCoV-229E), nsp15 has been found to be non-essential for viral replication in Mouse Hepatitis Virus [6] (MHV), SARS-CoV, and HCoV-229E. Some nsp15 mutations completely abolished RNA synthesis; however, these mutations resulted in misfolded and insoluble nsp15 when expressed in E. coli[6]. As a result, the loss of RNA synthesis is thought to be a knock-on effect on neighbouring polyprotein components that are critical for replication, as opposed to a genuine effect on viral replication through lack of nsp15 [6]. Further evidence of nsp15’s non-essential role in viral replication comes from insect nidoviruses and invertebrate roniviruses, which completely lack EndoU activity [8,9].
Although not essential for viral replication, recent studies suggest nsp15 plays a role in repressing activation of the host innate immune response [11–13]. During viral replication, positive-sense RNA is translated to produce the viral replication complex, which replicates the positive-sense RNA to produce negative-sense RNA. The negative-sense RNA then acts as a template to produce new positive-sense genomic RNA and subgenomic RNA. Subgenomic RNA consists of smaller transcribed sections of RNA produced by initiating transcription in the middle of the template strand (internal initiation), falling off the template strand before reaching the 5’ stop codon (premature termination), or by jumping off the template strand and reinitiating transcription further down the template (discontinuous transcription). This process produces short and long double-stranded RNA intermediates with polyuridine tracts at the 5′ end which can be recognized by pattern recognition receptors in the host cell such as RIG-I-like receptors (RLRs), protein kinase R (PKR), oligoadenylate synthases (OASes), and melanoma differentiation-associated gene 5 (MDA5). These sensors promote an innate and antiviral immune response [11,14,15] by activating the type I and III interferon (IFN) response, which induces expression of interferon -stimulated genes through the signal transducer and activator of transcription proteins 1 and 2 (STAT1/2) signaling pathways. By cleaving the 5′-polyuridine tracts in negative-sense viral RNA, nsp15, along with nsp16 and nsp10, limit the accumulation of MDA5-dependent pathogen-associated molecular patterns to delay the host’s immune response [16]. Loss of nsp15 activity has been shown to activate the interferon response and reduce viral titers in piglets infected with nsp15-deficient porcine epidemic diarrhea coronavirus (PEDV) [17] and mice infected with nsp15-deficient Mouse Hepatitis Virus [11]. It has also been demonstrated that nsp15 plays a role in disrupting formation of autophagosomes, which are double-membraned vesicles containing cellular material to be degraded.
Structural overview
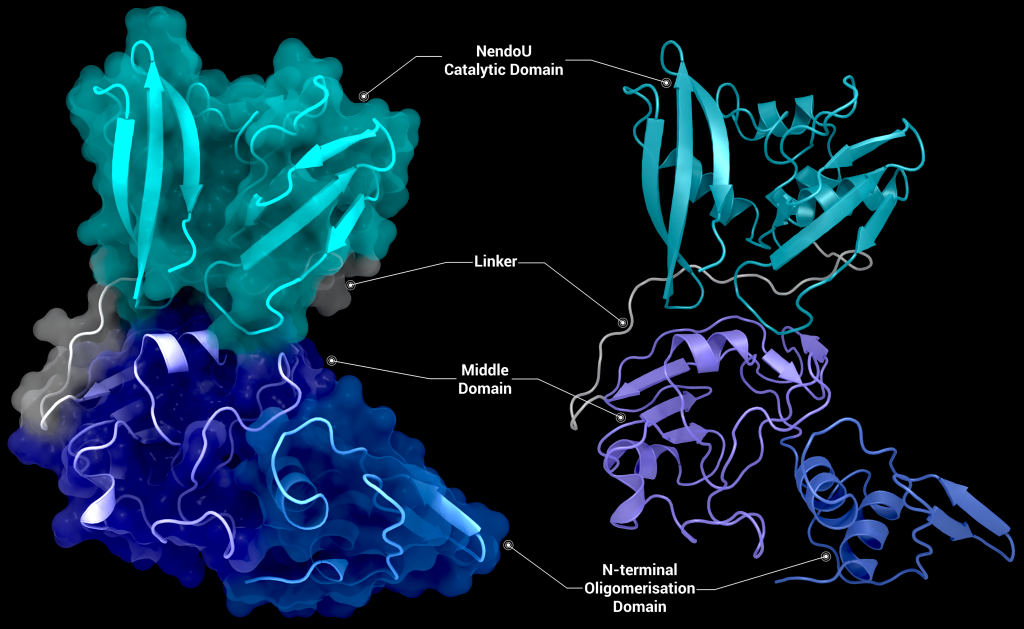
SARS-CoV-2 nsp15 consists of an N-terminal oligomerisation domain (Figure 2, blue), a middle domain (Figure 2, purple), and the catalytic C-terminal NendoU domain (Figure 2, teal). The Oligomerisation domain is formed from an anti-parallel β-sheet (β1-3) which wraps around helices α1 and α2. The middle domain consists of three β-hairpins (β5-6, β7-8, and β12-13), a mixed β-sheet (β4, β9, β10, β11, and β15), 2 α-helices (α3 and α4), and a right-handed 310 helix (η4). The catalytic NendoU domain contains two anti-parallel β-sheets (β16-18 and β19-21) which form a concave surface flanked by five α-helices (α6, α7, α8, α9, and α10). SARS-CoV-2 nsp15 shows high sequence identity with SARS-CoV nsp15 (88%) and lower sequence identity with MERS-CoV (51%), however the overall structural similarity is very high between the three viruses [1]. Three structures have been solved for SARS-CoV nsp15 (PDB entries 2H85 [18], 2OZK [19], and 2RHB [20]) one structure of MERS nsp15 (PDBID: 5YVD [21]), two structures from mouse hepatitis virus (2GTH and 2GTI [3]), and one structure from human coronavirus 229E (PDB entry 4S1T). As of writing this review 20 structures of SARS-CoV-2 nsp15 have been solved with a variety of bound ligands using X-ray crystallography and cryo-EM (Table 1) [1,22,23].
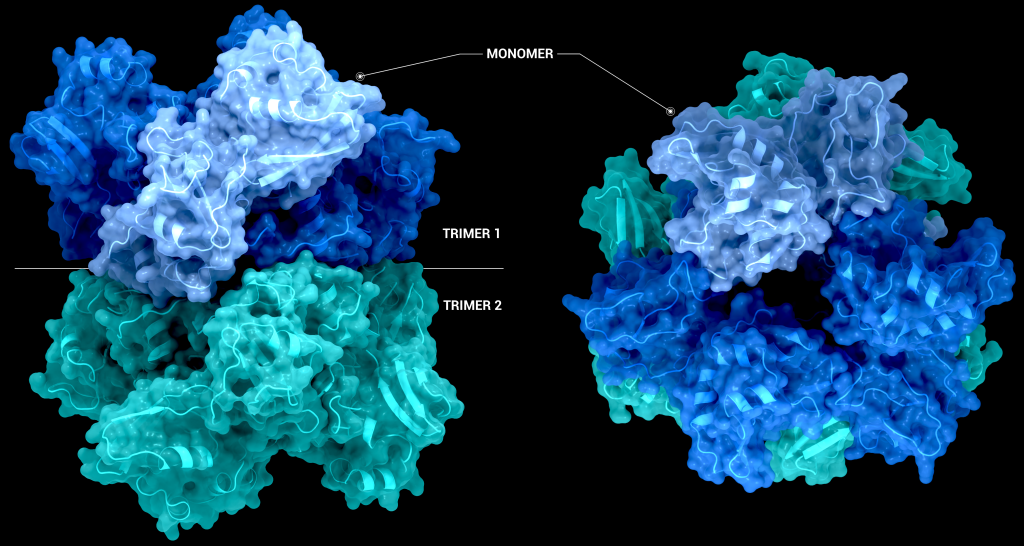
The biological assembly of nsp15 is a double-ringed hexamer made up of a dimer of trimers (point group 32, Figure 3). The trimeric form retains some ribonuclease activity, but the monomer presents with only residual cleavage [24] The hexamer is stabilised by an N-terminal oligomerisation domain present in each monomer. A crystal structure from SARS-CoV with a 28 amino acid N-terminal truncation (PDB entry 2H85) presented with a misfolded endoU active site, suggesting oligomerisation may act as an allosteric activation switch [19]. The six monomers come together to form the active enzyme with a 100 Å long negatively charged channel 10 to 15 Å wide open to solvent at the top, bottom, and on three separate side openings in the middle of the hexamer. Formation of the hexamer is essential for enzymatic activity, making the oligomerisation interfaces a potential target for structure-based drug design.
The active site of nsp15 is an electropositive pocket which lies at the interface between each monomer’s NendoU domain. The active site is highly conserved between SARS-CoV-2, SARS-CoV, and MERS proteins. Six key residues (His235, His250, Lys290, Thr341, Tyr343, and Ser294) are arranged in a shallow groove in the N-terminal NendoU domain [1]. His235, His250, and Lys290 are proposed to act as a catalytic triad, using a similar mechanism to that observed in RNase A [23]. However, RNase A is metal-independent, while SARS-CoV-2 nsp15 is Mn2+ dependent, so the mechanism is not an exact match. Mutation of either histidine in the catalytic triad to alanine eliminates RNA cleavage activity in nsp15 but has no effect on the formation of stable hexamers, showing they are not a factor in nsp15 oligomerisation [22]. This is unsurprising, as the N-terminal oligomerisation domain is the key player in the formation of the hexamer, but formation of the hexamer clearly plays an allosteric role in the formation of the active site, as activity in the monomer is significantly reduced.
Uracil specificity is proposed to be governed by Ser294 [20], with the main chain nitrogen of Ser294 predicted to interact with the carbonyl O2 oxygen of uracil and the hydroxyl group of Ser294 binding to uracil N3. However, mutation studies on homologs have shown that a Ser294Ala mutation significantly decreased activity without completely abolishing it[18] and negates uridine specificity. Tyr343 is likely important in governing uracil specificity, as shown by van der Waals stacking between the ribose sugar or Uridine and Tyr343 in cryo-EM structures [20,21]. Mutation of Tyr343 equivalent residues in SARS-CoV and MERS to alanine caused near complete loss of nuclease activity [20,21], suggesting a key role in enzymatic activity.
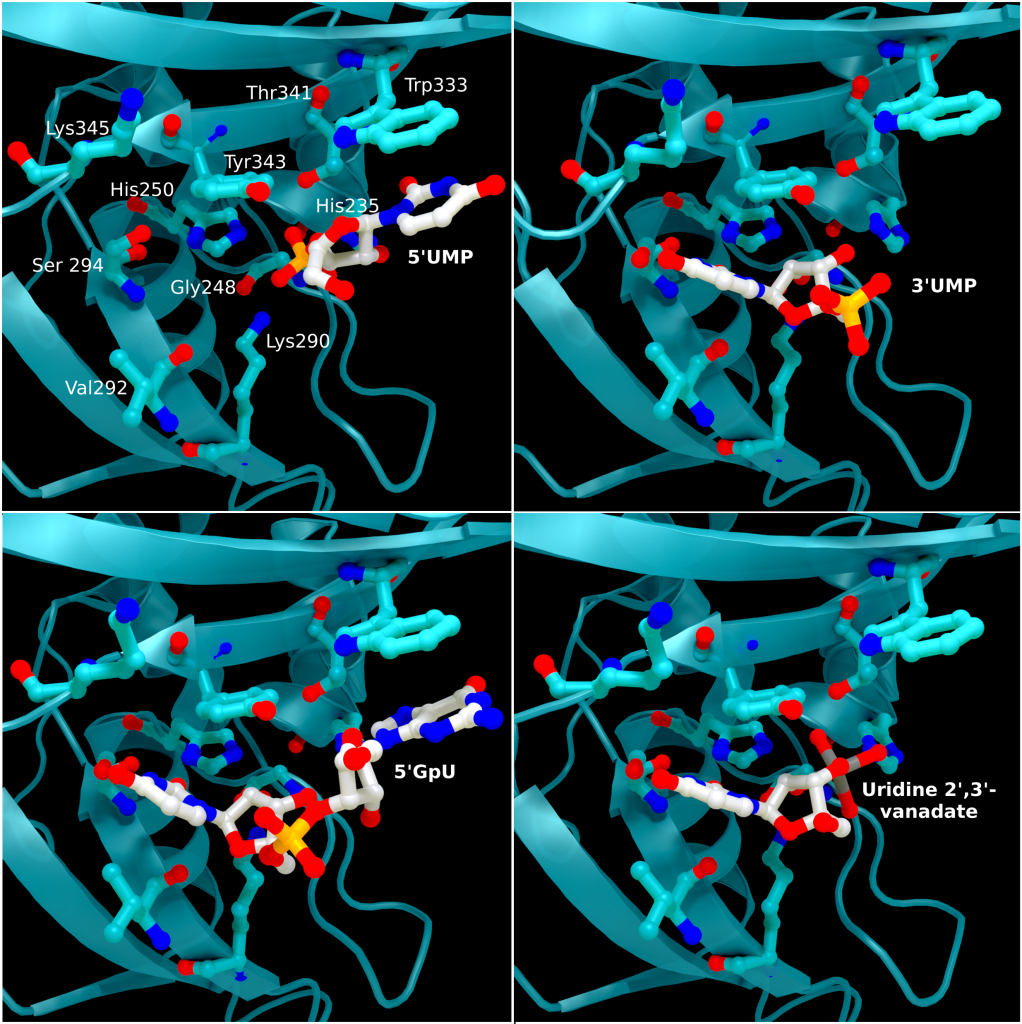
The structure of SARS-CoV-2 nsp15 has been solved in the presence of various catalytic intermediates, including 5′UMP (PDB entry: 6WLC), 3′UMP (PDB entry: 6X4I), 5′GpU (PDB entry: 6X1B), and uridine 2′,3′-vanadate (PDB entry: 7K1L). All intermediates bound to the C-terminal catalytic domain, interacting with the seven conserved active site residues (His235, His250, Lys290, Trp333, Thr341, Tyr343, Ser294, Gly248, Lys345, and Val292) and the structures showed no significant conformational deviations from each other (Cα RMSD = 0.29 Å). The uracil moiety of 5′UMP, guanylyl(3’-5’)uridine (GpU), and uridine 2′,3′-vanadate are all bound by Ser294 and Leu346 (Figure 4 Top left, bottom left, and bottom right, respectively), reinforcing the idea of uracil recognition being mediated by these residues. The combination of these structures confirms the predicted parallels between the reaction mechanism of SARS-CoV-2 nsp15 and RNAse A. The 5′UMP, 5′GpU, and uridine 2′-,3′-vanadate bound structures support the previously proposed hypothesis about uracil and purine base discrimination with Ser294 playing a key role [23]. Contrary to this finding, the 3′UMP bound structure shows the uracil base forming a stacking interaction with Trp333 (Figure 4, top right), the guanine binding site identified in the 5′GpU complex, suggesting nsp15’s active site can accommodate both purine and pyrimidine bases. However, the Trp333 interacting base is likely less relevant when binding larger RNA molecules as it provides a potential stacking interaction for bases without selectivity [23].
Comparison of these ligand-bound structures with RNase A catalytic sites suggests nsp15 acts through a similar reaction mechanism [23]. Based on these findings a two-step mechanism has been proposed starting with a transphosphorylation reaction whereby His250 acts as a base and deprotonates 2′OH of the RNA ribose, with Lys290 stabilising the negative charge that builds up during the transition state. His235 then acts as a general acid donating a proton for the departing 5′OH group. This is followed by a hydrolysis step where the roles of His250 and His235 are reversed, with His235 deprotonating a water molecule and His250 acing as a proton donor for the 5′OH leaving group to convert the 2′-3′ cyclic phosphate back to 2′OH and a 3′-phosphoryl group. Despite the similar mechanisms, the structural environments of His235 in nsp15 and the RNase A equivalent (His119) differ significantly, with the residues being ~8 Å apart and making several different hydrogen bonding interactions. These differences may provide an answer as to why nsp15 is much more sensitive to pH change compared to Rnase A [22]. What remains unclear is the contribution of Mn2+ to the reaction mechanism, particularly as an Mn2+ binding site has not been located in SARS-CoV-2 nsp15 [22].
Therapeutic interest of the protein
As previously mentioned, knockout studies on nsp15 have shown it is not essential for viral replication. Despite this, a nsp15 inhibitor could provide an effective treatment against SARS-CoV-2 by hampering its evasion and modulation of the innate immune response to help promote longer-lasting immunity. Targeting nsp15 is particularly interesting as nsp15 has no close human homologues [25], thereby potentially reducing harmful side effects. A number of biochemical assays have been performed on nsp15 to screen previously approved drugs and various libraries for inhibition of nsp15, as well as a number of in-silico studies to dock approved therapeutics to guide drug design efforts. A fragment screening study has also been performed that yielded 6 small molecule fragments.
Benzopurpurin B, C-473872 (CAS registry number: 331675-78-6), and Congo Red, as well as small molecular Rnase A inhibitors, have been shown to inhibit nsp15 activity and reduce infectivity of SARS-CoV in Vero cells [26] but further testing on SARS-CoV-2 nsp15 is required. Additionally, nsp15 has been screened against the ReFrame [27], Pandemic Response Box (Medicines for Malaria Venture (MMV) & Drugs for neglected disease initiative (DNDi)), and Covid Box drug repurposing libraries for 50% inhibition below concentration of 10 µM, identifying 23, 1, and 0 hits respectively from the libraries [25]. Two fluorescence resonance energy transfer (FRET) assays to determine the half-maximal inhibitory concentration (IC50) reduced the hits to 12 (11 in ReFrame, 1 in Pandemic Response Box), which were whittled down to 3 (Exebryl-1, Piroxantrone, and MMV1580853) after 9 were identified as false positives due to the production of reactive oxygen species such as H2O2, which destabilized protein in the assay. Ligand binding was assessed using high resolution mass spectrometry. Piroxantrone and MMV1580853 showed significantly weaker binding and ultimately no antiviral activity in SARS-CoV-2 assays. Exebryl-1 bound with an affinity constant Kd of ~12 µM per monomer in the first instance, with approximately four molecules binding to one monomer on average per 100 µM Exebryl-1; and molecular docking of Exebryl-1 against PDB entry 6XDH using an automated Qvina docking workflow [28] showed binding in a pocket close to and within the active site. Exebryl-1 demonstrated antiviral activity in three separate assays at concentrations over 10 µM. However, based on blood plasma levels in Sprague-Dawley rats after an oral dose of 100 mg/kg reaching only 9 µM after 1 hour, and dropping to 4 µM after 4 hours, Exebryl-1 is not expected to reach therapeutic levels in its current state [25].
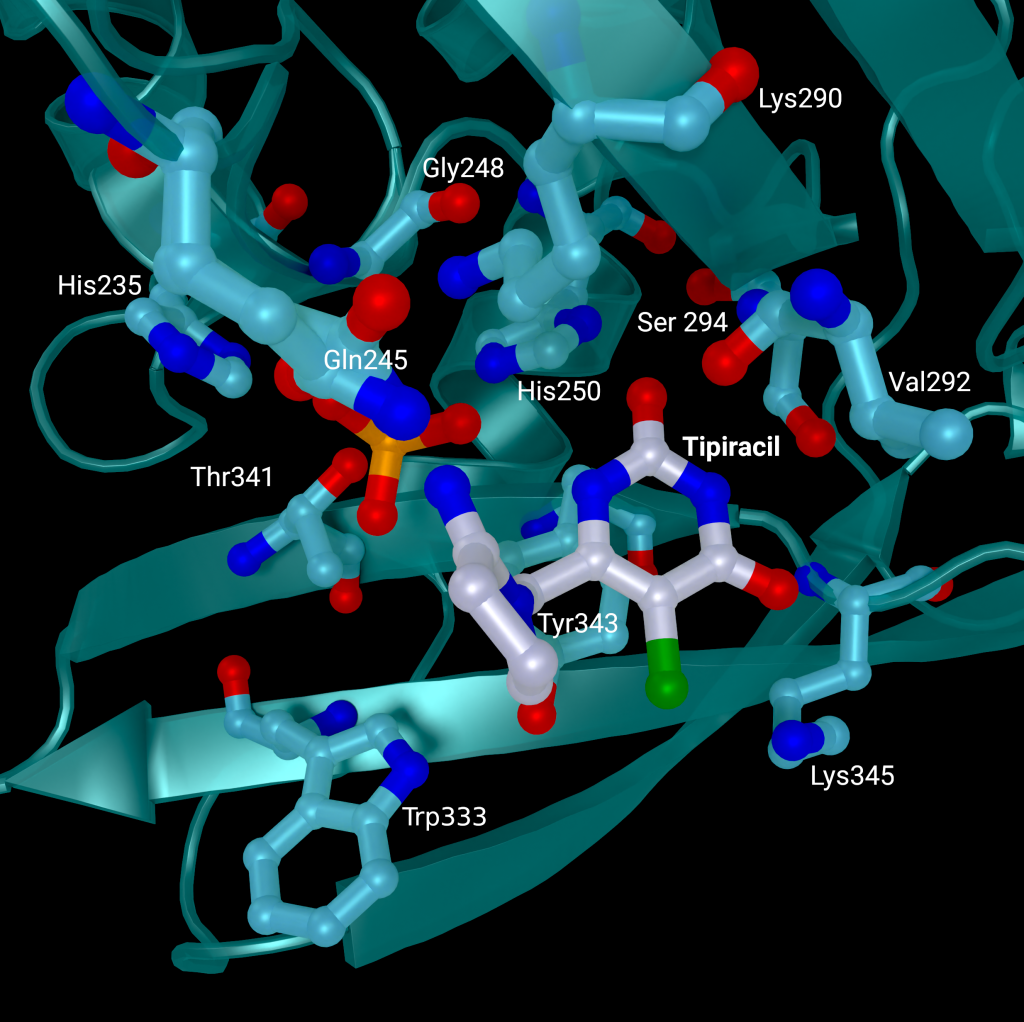
A repurposed colorectal cancer drug, Tipiracil, has been found to partially inhibit nsp15 activity in biochemical assays. However, the efficacy is greatly decreased in the presence of increased Mn2+ concentrations. A structure of nsp15 with Tipiracil interacting with the uridine binding pocket has also been solved (PDB entry: 6WXC), with its uracil ring stacking against Tyr341 and forming several hydrogen bonds with Ser294, Lys345, and His250 (Figure 5) as well as several interactions with other active site residues through water and phosphate molecules. The only unique interaction for this ligand is between the Iminopyrrolidin nitrogen of Tipiracil and Gln245 (Figure 5). Although not an immediate treatment option, the uracil derivative drug provides a potential scaffold for further SARS-CoV-2 nsp15 inhibitor development [23]. Based on Tipiracil binding at the active site a library of 85 flavinoid compounds were docked using the molecular mechanics/generalized Born surface area (MMGBSA) method and molecular dynamics with nsp15 (PDB entry 6WXC) as part of an in-silico study; but binding was found to be significantly weaker than Tipiracil in all cases [29].
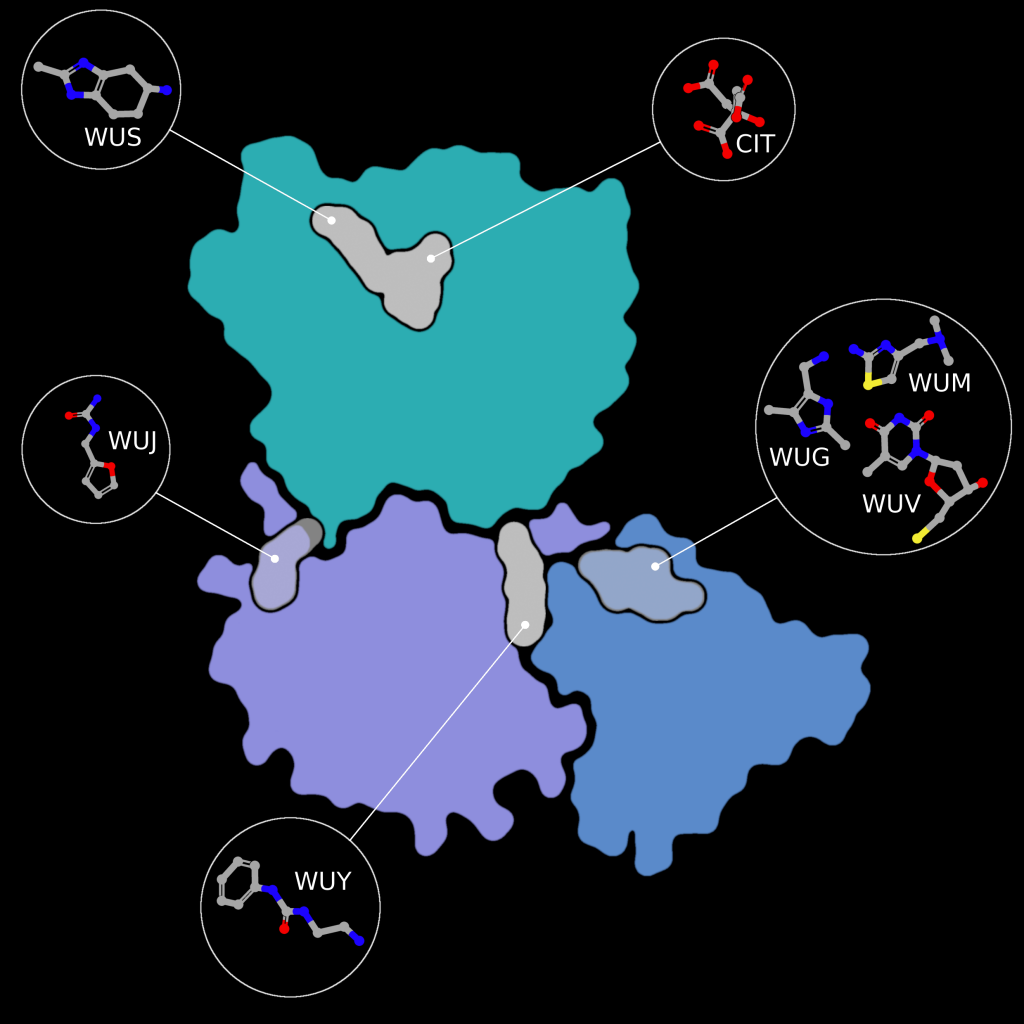
Fragment screens have been performed on nsp15, with six structures currently available in the PDB without an accompanying publication. In addition to the soaked fragments present in these structures, all show a citrate molecule bound to the catalytic NendouU domain (Figure 6, CIT), with one fragment bound adjacent to citrate (PDB entry 5S70, Figure 6, EN300-181428 (WUS)) through a stacking interaction with Trp333 and a hydrogen bond between the NO3 hydrogen of EN300-181428 and O5 of the citrate molecule. Four fragments are bound at the interface between the middle domain (Figure 6, purple) and the N-terminal oligomerisation domain (Figure 6, blue), including FUZS-5 (PDB entry 5S71, Figure 6, WUV) Z2889976755 (PDB entry 5S6X, Figure 6, WUG), BBL029427 (PDB entry 5S72, Figure 6, WUY), and PB2255187532 (PDB entry 5S6Z, Figure 6, WUM). Finally, BBL029427 (PDB entry 5S6Y, Figure 6, WUJ) is bound to a loop connecting beta strands in the middle domain. Unfortunately, the crystal packing in these structures prevents the formation of the active double-ringed hexamer structure using symmetry related molecules, making it difficult to assess how the fragments interact with the active hexamer. However, this monomeric crystal form could provide a starting point for the design of a drug to break up formation of the active hexamer by interfering with surfaces on the N-terminal oligomerization domain.
Molecular docking, all-atom molecular dynamics, and an assessment of absorption, distribution, metabolism, and excretion (ADME) properties have been carried out on PDB entry 6W01 using 15 scalarane sesterterpenes, compounds purified from Red Sea marine sponges with a variety of relevant pharmacological activities. to assess their efficacy as drug targets to inhibit nsp15 [30]. Eight compounds were found to have equivalent or better binding energies compared to the reference ligand, Benzopurpurin 4B. All eight compounds bound the C-terminal catalytic domain in the large shallow active site, forming polar interactions with the catalytic triad (His235, His250, and Lys290), interacting with Trp333 through π-stacking, and forming at least one hydrogen bond with Lys290 and further anchoring hydrogen bonds with Gly248 and/or Gln245 [30]. Two of the eight were used in all atom molecular dynamics simulations and showed good stability, high negative binding free energies, and scored well on ADME drug property predictions.
In-silico docking investigations of 32 phytochemicals from Asparagus racemous have also been performed on nsp15 (PDBID: 6W01). The top 5 ligands (Asparoside-C, Asparoside-F, Rutin, Asparoside-D, and Racemoside-A) bound at the C-terminal active site with binding free energy scores between ‒7.165 kcal/mol and ‒5.993 kcal/mol. Complexes of nsp15 and Asparoside-C, -F, and -D were subjected to further analysis by 100 ns molecular dynamics simulations, which found Asparoside-D and -F to have favorable binding interactions and better affinity than the control ligand Remdesivir [31]. 23 previously approved drugs have also been docked to nsp15, with three demonstrating high predicted binding affinities between ‒9.1 and ‒9.6 kcal/mol (Saquinavir, Aprepitant, and Valrubicin) [32]. However, the pocket Saquinavir, Aprepitant, and Valrubicin are docked to sites on the opposite side of the active site pocket which houses the catalytic triad, approximately 17 Å away. Barring an undetermined allosteric effect caused by this binding, which the paper makes no mention of, further development of these drug targets “…modifying them to fit to the SARS-CoV-2 nsp15 active site pocket precisely” needs to be rethought as the active site has not been targeted in the first instance.
Complementary knowledge
The enzymatic activity of nsp15 and its crystal structure have been demonstrated, but the exact role in viral replication remains unclear. SARS-CoV nsp15 has been shown to co-localize with replicating RNA [33] around the nucleus as well as nsp8 and nsp12 from the replication/transcription complex in in situ studies [34], in the presence and absence of RNA. It was also shown that SARS-CoV nsp15 does not co-localise with the M protein [34]. Yeast two-hybrid screens and glutathione S-transferase (GST) pulldown assays have also identified nsp8 and nsp12 as potential binding partners to SARS-CoV nsp15 [35].
Furthermore, nsp15 has demonstrated a strong inhibitory effect on interferon (IFN) production and interferon regulatory factor 3 nuclear localization in in-vitro co-expression assays against the Cantell strain of Sendai virus with nsp13, nsp14, and accessory protein ORF6 [36]. However, interferon antagonization in in-vitro conditions is not necessarily representative of real infection, individual protein expression levels can vary greatly compared to overexpression studies and altered localization can have a significant effect[36]. The individual contribution or mechanism of nsp15 interferon inhibition is not discussed by Yuen et al 2020 in this study. Overall SARS-CoV-2 appears less effective at suppressing interferon signaling compared to SARS-CoV due to the loss of SARS-CoV-2 papain-like protease (PLpro) as an interferon antagonist [36]. Reverse genetic studies (analysis of a resulting phenotype following genetic engineering) have suggested that ORF6 is the major player in interferon suppression instead [37]. However, SARS-CoV-2 ORF6 is also less conserved between SARS-CoV and SARS-CoV-2 at only 69% sequence identity and only 4 of 10 key amino acids identified from SARS-CoV ORF6 being present in SARS-CoV-2 ORF6 [36].
It has been shown that nsp15 activity is highly dependent on the presence of Mn2+ ions, showing greatly reduced activity in the presence of Mg2+ ions. In the presence of Mn2+ nsp15 was able to cleave all four uridine sites in an eicosamer, a 20-subunit oligomer consisting of 5′GAACU↓CAU↓GGACCU↓U↓GGCAG3′, with no preference for sequence and increased cleavage rate with rising metal ion concentration [23]. This is particularly interesting as Mn2+ enhances activity in SARS-CoV nsp15, but protein activity does not depend on the presence of Mn2+, and no metal binding sites have been identified in coronavirus structures to date [18]. Considering SARS-CoV-2 nsp15 shares 88% sequence identity with SARS-CoV nsp15, and all active site residues are conserved, SARS-CoV 2 nsp15’s dependence on Mn2+ is a significant difference between the enzymes. Further to this, nsp15 alone is promiscuous, cutting any uridine sites in RNA, but becomes site-specific when in complex with nsp8 and nsp12 and leaves uridine tails between 5 and 10 bases long [16].
A library of 5000 small molecule compounds has been screened against nsp15 for inhibition of nuclease activity, with twelve compounds showing potential as antiviral treatments in a fluorescent biochemical kinetic screen. Further analysis using a gel-based assay found only one compound, NSC95397, able to inhibit nuclease activity at a concentration of 10 µM. However, tests on SARS-CoV 2 infected VERO E6 cells found the compound toxic at concentrations above 10 µM and ineffective at inhibiting viral growth at lower concentrations [38].
A fluorescence resonance energy transfer (FRET) assay has been performed to measure nsp15 activity on a 6-mer oligonucleotide (5′-AAAUAA) with a 5′-fluorescein and 3′-TAMRA label [21,22]. Activity is measured through an increase in fluorescence caused by the removal of the 5′-TAMRA label. Nsp15 activity was confirmed for the wild-type protein and abolished in H235A and H250A mutants [22]. FRET analysis was paired with liquid chromatography electrospray ionization mass spectrometry to demonstrate that nsp15 3′RNA products show a preference for accumulation of 2′-3′ cyclic phosphate (80%) compared to 3′-phosphate, a significant difference compared to RNAse A which generates a 2’-3’ cyclic phosphate which is then hydrolysed to a 3’-phosphate.
Summary
SARS-CoV 2 nsp15 is an RNA uridylate‐specific Mn2+-dependent [3] endoribonuclease from the nidoviral endoU (NendoU) family, which acts on single-stranded and double-stranded RNA to help SARS-CoV-2 evade detection by the innate immune response. Knockout studies have demonstrated that nsp15 is not essential for viral replication, but numerous studies have shown a reduction in viral titre and virulence in nsp15-deficient SARS-CoV-2 when studied in the presence of an effective immune response.
The sequence of nsp15 is highly conserved between SARS-CoV-2, SARS-CoV, MERS-CoV, and HCoV-229E, as is the fold of the monomer and active hexamer. The monomer consists of three domains, the N-terminal oligomerisation domain, a middle domain, and the NendoU catalytic domain which houses the active site. The active site is a shallow groove made up of six key residues (His235, His250, Lys290, Thr341, Tyr343, and Ser294). A series of structures with different catalytic intermediates have been solved and the reaction mechanism is predicted to act in a similar manner to the well-studied RNaseA enzyme. However, nsp15’s dependence on manganese, where RNase A’s activity is metal independent, throws some aspersions on this theory.
Three in-silico drug screeningstudies have been performed on nsp15, two using 6W01 and one using 6WXC as the protein models. 6W01 is a citrate bound nsp15 structure solved to 1.9 Å resolution, with acceptable data processing and refinement statistics overall, the only minor concern is that 5% of the residues in both chains show one issue with their geometry, and a small subset of that 5% show an issue in their fit to the electron density. 6WXC is a Tipiracil bound nsp15 structure solved to 1.85 Å resolution, it faces a similar minor problem to 6W01 with 7% of residues in both chains showing one issue with their geometry but with fewer electron density fit outliers. Use of either model should present no major stumbling blocks for simulation studies.
Discussion & Outlook
Nsp15 has been one of the lesser explored proteins compared to other SARS-CoV 2 proteins, such as the main protease and the papain-like protease, which have undergone extensive in-silico drug design studies through a number of large collaborative efforts between universities, synchrotrons, and other organizations [39–45] to feed into the COVID Moonshot project [46]. Overall, the structural work on nsp15 has been sound and all available models could provide a good starting structure for computational drug design. A series of structures with catalytic intermediates suggests a mechanism akin to RNase A, however, the dependence of nsp15 on Mn2+ suggests a departure from this mechanism as RNase A’s mechanism is metal independent. Follow up in-silico studies (described above) were based on well validated models with acceptable statistics for the resolution the structures were solved at, although none have yet pointed to a viable lead compound for clinical application. Nsp15 not being essential for viral replication makes it a much less desirable target for structure-based drug design compared to other essential viral proteins. However, the impact of nsp15 on SARS-CoV-2’s virulence by repressing the innate immune response shows a potential avenue to weaken SARS-CoV-2 through inhibition of nsp15 to allow the immune system to fight off infection before it becomes more severe.
Acknowledgements
This work was supported by the German Federal Ministry of Education and Research [grant no. 05K19WWA], Deutsche Forschungsgemeinschaft [project TH2135/2-1]. The authors would also like to thank Johannes Kaub and Rosemary Wilson for support and discussion. All figures are courtesy of the Coronavirus Structural Task Force (insidecorona.net), who retains copyright for the text and the figures..
[1] Kim Y, Jedrzejczak R, Maltseva NI, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Science. 2020;29:1596–1605.
[2] Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192.
[3] Ivanov KA, Hertzig T, Rozanov M, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci U S A. 2004;101:12694–12699.
[4] Naqvi AAT, Fatima K, Mohammad T, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165878.
[5] Bhardwaj K, Sun J, Holzenburg A, et al. RNA Recognition and Cleavage by the SARS Coronavirus Endoribonuclease. J Mol Biol. 2006;361:243–256.
[6] Deng X, Baker SC. An “Old” protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses. Virology. 2018;517:157–163.
[7] Snijder EJ, Decroly E, Ziebuhr J. Chapter Three - The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. In: Ziebuhr J, editor. Advances in Virus Research [Internet]. Academic Press; 2016 [cited 2022 Jan 7]. p. 59–126. Available from: https://www.sciencedirect.com/science/article/pii/S0065352716300471.
[8] Nga PT, Parquet M del C, Lauber C, et al. Discovery of the First Insect Nidovirus, a Missing Evolutionary Link in the Emergence of the Largest RNA Virus Genomes. PLOS Pathogens. 2011;7:e1002215.
[9] Lauber C, Ziebuhr J, Junglen S, et al. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch Virol. 2012;157:1623–1628.
[10] Tomasello G, Armenia I, Molla G. The Protein Imager: a full-featured online molecular viewer interface with server-side HQ-rendering capabilities. Bioinformatics. 2020;36:2909–2911.
[11] Deng X, Hackbart M, Mettelman RC, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. PNAS. 2017;114:E4251–E4260.
[12] Kindler E, Gil-Cruz C, Spanier J, et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLOS Pathogens. 2017;13:e1006195.
[13] Volk A, Hackbart M, Deng X, et al. Coronavirus Endoribonuclease and Deubiquitinating Interferon Antagonists Differentially Modulate the Host Response during Replication in Macrophages. Journal of Virology [Internet]. 2020 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/JVI.00178-20.
[14] Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105.
[15] Mandilara G, Koutsi MA, Agelopoulos M, et al. The Role of Coronavirus RNA-Processing Enzymes in Innate Immune Evasion. Life (Basel). 2021;11:571.
[16] Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci U S A. 2020;117:8094–8103.
[17] Deng X, Geelen A van, Buckley AC, et al. Coronavirus Endoribonuclease Activity in Porcine Epidemic Diarrhea Virus Suppresses Type I and Type III Interferon Responses. Journal of Virology [Internet]. 2019 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/JVI.02000-18.
[18] Ricagno S, Egloff M-P, Ulferts R, et al. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. PNAS. 2006;103:11892–11897.
[19] Joseph JS, Saikatendu KS, Subramanian V, et al. Crystal Structure of a Monomeric Form of Severe Acute Respiratory Syndrome Coronavirus Endonuclease nsp15 Suggests a Role for Hexamerization as an Allosteric Switch. Journal of Virology [Internet]. 2007 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/JVI.02817-06.
[20] Bhardwaj K, Palaninathan S, Alcantara JMO, et al. Structural and Functional Analyses of the Severe Acute Respiratory Syndrome Coronavirus Endoribonuclease Nsp15*. Journal of Biological Chemistry. 2008;283:3655–3664.
[21] Zhang L, Li L, Yan L, et al. Structural and Biochemical Characterization of Endoribonuclease Nsp15 Encoded by Middle East Respiratory Syndrome Coronavirus. Journal of Virology [Internet]. 2018 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/JVI.00893-18.
[22] Pillon MC, Frazier MN, Dillard LB, et al. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat Commun. 2021;12:636.
[23] Kim Y, Wower J, Maltseva N, et al. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun Biol. 2021;4:1–11.
[24] Saramago M, Costa VG, Souza CS, et al. The nsp15 Nuclease as a Good Target to Combat SARS-CoV-2: Mechanism of Action and Its Inactivation with FDA-Approved Drugs. Microorganisms. 2022;10:342.
[25] Choi R, Zhou M, Shek R, et al. High-throughput screening of the ReFRAME, Pandemic Box, and COVID Box drug repurposing libraries against SARS-CoV-2 nsp15 endoribonuclease to identify small-molecule inhibitors of viral activity. PLOS ONE. 2021;16:e0250019.
[26] Ortiz-Alcantara J, Bhardwaj K, Palaninathan S, et al. Small molecule inhibitors of the SARS-CoV Nsp15 endoribonuclease. Virus Adaptation and Treatment. 2010;2:125–133.
[27] Janes J, Young ME, Chen E, et al. The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. PNAS. 2018;115:10750–10755.
[28] Alhossary A, Handoko SD, Mu Y, et al. Fast, accurate, and reliable molecular docking with QuickVina 2. Bioinformatics. 2015;31:2214–2216.
[29] Mishra GP, Bhadane RN, Panigrahi D, et al. The interaction of the bioflavonoids with five SARS-CoV-2 proteins targets: An in silico study. Comput Biol Med. 2021;134:104464.
[30] Elhady SS, Abdelhameed RFA, Malatani RT, et al. Molecular Docking and Dynamics Simulation Study of Hyrtios erectus Isolated Scalarane Sesterterpenes as Potential SARS-CoV-2 Dual Target Inhibitors. Biology (Basel). 2021;10:389.
[31] Chikhale RV, Sinha SK, Patil RB, et al. In-silico investigation of phytochemicals from Asparagus racemosus as plausible antiviral agent in COVID-19. J Biomol Struct Dyn. 2021;39:5033–5047.
[32] Mahmud S, Elfiky AA, Amin A, et al. Targeting SARS-CoV-2 nonstructural protein 15 endoribonuclease: an in silico perspective. Future Virol. :10.2217/fvl-2020–0233.
[33] Shi ST, Schiller JJ, Kanjanahaluethai A, et al. Colocalization and Membrane Association of Murine Hepatitis Virus Gene 1 Products and De Novo-Synthesized Viral RNA in Infected Cells. Journal of Virology [Internet]. 1999 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/JVI.73.7.5957-5969.1999.
[34] Athmer J, Fehr AR, Grunewald M, et al. In Situ Tagged nsp15 Reveals Interactions with Coronavirus Replication/Transcription Complex-Associated Proteins. mBio [Internet]. 2017 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/mBio.02320-16.
[35] Imbert I, Snijder EJ, Dimitrova M, et al. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148.
[36] Yuen C-K, Lam J-Y, Wong W-M, et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 9:1418–1428.
[37] Schroeder S, Pott F, Niemeyer D, et al. Interferon antagonism by SARS-CoV-2: a functional study using reverse genetics. The Lancet Microbe. 2021;2:e210–e218.
[38] Canal B, Fujisawa R, McClure AW, et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp15 endoribonuclease. Biochem J. 2021;478:2465–2479.
[39] Cantrelle F-X, Boll E, Brier L, et al. NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment. Angewandte Chemie International Edition. 2021;60:25428–25435.
[40] Newman JA, Douangamath A, Yadzani S, et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat Commun. 2021;12:4848.
[41] Zhao Y, Du X, Duan Y, et al. High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors. Protein Cell. 2021;12:877–888.
[42] Ma C, Sacco MD, Xia Z, et al. Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay. ACS Cent Sci. 2021;7:1245–1260.
[43] Douangamath A, Fearon D, Gehrtz P, et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat Commun. 2020;11:5047.
[44] Ahmad S, Abdullah I, Lee YK, et al. Extensive Crystallographic Fragment-Based Approach to Design SARS CoV2 3CLpro Main Protease Inhibitors and Related Metadata. 2021 [cited 2022 Jan 6]; Available from: https://chemrxiv.org/engage/chemrxiv/article-details/60c753c8469df4c73ef44e13.
[45] Günther S, Reinke PYA, Fernández-García Y, et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372:642–646.
[46] Consortium TCM, Achdout H, Aimon A, et al. Open Science Discovery of Oral Non-Covalent SARS-CoV-2 Main Protease Inhibitor Therapeutics [Internet]. 2021 [cited 2022 Jan 6]. p. 2020.10.29.339317. Available from: https://www.biorxiv.org/content/10.1101/2020.10.29.339317v2.
Für diesen Beitrag exisitiert leider keine deutsche Übersetzung.
Since the outbreak of SARS-CoV-2, infection has continued to spread. At the same time, governmental agencies around the world have adjusted the rules to prevent its spread. Information sources as basis for these rules have been obtained from scientific studies, public health research and simulation tests to understand the efficiency of mask types in preventing spread of infection by SARS-CoV-2. In this article, we will look at the mask types in use today, how much they can impede viral droplets and aerosols and how the construction of different masks helps to protect us from infection by SARS-CoV-2.
SARS-CoV-2 droplet sizes and viral transmission
The SARS-CoV-2 virus can be transmitted via droplets and aerosols.
Droplets are particles of sizes varying from 0.05 to 500 μm. They are directly emitted while breathing or talking. After being released into the air, larger droplets fall to the ground and others rapidly evaporate to form droplet nuclei less than 5 µm of size, also called aerosols, containing viruses in the range of 0.02 to 0.3 μm. Droplet nuclei can remain suspended in air for a longer time compared to large droplets and potentially contribute to airborne transmission1,2,3.
SARS-CoV-2 has been observed to be transmitted via 3 modes:4,5,6
- Contact transmission (usually via direct contact with infected persons, surfaces, or air)
- Droplet transmission over short distances when a person is close to an infected person
- Aerosol transmission over longer distances via inhalation of aerosols that remain airborne and travel with the air
Although maintaining a safe distance from an infected or possibly infected person will prevent viral spread via direct contact and droplet transmission, maintaining a safe distance may not be able to prevent spread of infection through airborne aerosols. This is why it becomes even more important to wear a mask.
Mask types and structure
Surgical masks, also called medical face masks or mouth-nose protection (MNS), are disposable products that are normally used in clinics or in doctor's offices on a daily basis. They are made of special plastics with multiple layers. They have a rectangular shape with wrinkles so that the mask can adapt to the face. The front (outside) is often coloured, the back (inside) is not. The masks have ear loops and a wire noseband (see Figure 1).
Due to the shape and fit of most medical face masks, some of the breathing air can flow past the edges. Especially during inhalation, unfiltered breathing air can be sucked in. Therefore, medical face masks usually offer the wearer less protection against pathogenic aerosols than particle-filtering half-masks (FFP). Medical face masks, however, can protect the mouth and nose of the wearer from pathogen transmission via direct contact, for example with contaminated hands.
Since they are medical devices, their manufacturing and distribution must be carried out in accordance with medical device law. They must therefore comply with the legal requirements and the European standard EN 14683:2019-10. Only then can manufacturers mark the medical masks with the CE mark and distribute them freely in Europe. This is subject to supervision by competent authorities7.
Particle filtering half masks / filtering facepieces (FFP) are objects of personal protective equipment (PPE) within the framework of occupational health and safety. They protect the wearer of the mask from particles, droplets, and aerosols. When worn correctly, FFP masks are tightly attached and offer external and self-protection. Since the masks are disposable products as intended by the manufacturer, they should be changed regularly and disposed of after use.
FFP masks are produced either with or without an exhalation valve. Masks without exhalation valve filter both the inhaled air and the exhaled air over the mask surface and therefore offer both self-protection and external protection. Masks with valves offer less external protection because exhaled aerosols are not intercepted by the filter material but are only slowed down and swirled to a certain extent by the valve.
Like medical face masks, FFP masks must comply with clear requirements of laws and technical standards. In particular, the filter performance of the mask material is tested with aerosols in accordance with the European standard EN 149:2001+A1:2009. FFP2 masks must filter at least 94% of the test aerosols, for FFP3 masks the minimum is even 99% . They are therefore proven to provide effective protection against aerosols. The test standard, together with the CE mark and the four-digit identification number of the notified body, is printed on the surface of the FFP mask7.

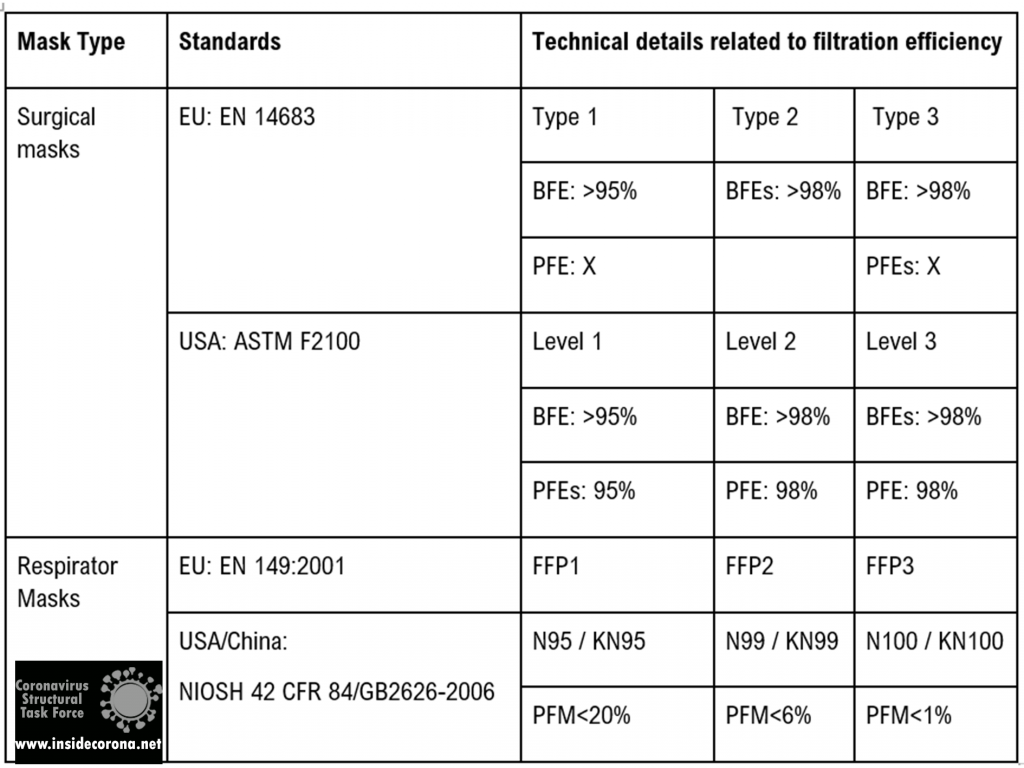
Mask standards
The table below shows the currently accepted standards for masks and how they are effective in filtering out bacteria as well as particles.
Mechanisms of protection
Masks ensure protection from viral spread in three main ways1,5:
Flow resistance inhibits the momentum of exhaled droplets and the velocity of incoming airborne aerosols. This significantly reduces the risk of infection in the vicinity of an infected person, protecting third parties as well. This is afforded by surgical masks, FFP2/N95/KN95, or better particle filtering respirator masks.
Droplet filtration blocks out large droplets via gravity sedimentation, inertial impaction, and minimizing contact of hand to mouth, nose, or other facial canals with access to the respiratory tract. It is afforded by most kinds of masks.
Aerosol filtration reduces the spread of aerosols via interception, diffusion, and electrostatic attraction. Electrostatic effects likely result in charge transfer with nanoscale aerosol particles. It is afforded by FFP2/N95/KN95 or better particle filtering respirator masks.
At small aerosol droplet sizes in the range of 0.1 to 1 μm, the mask layers prevent particles from passing mainly by blocking movement of particles with the fibers in the filter layer and, hence, not allowing diffusion. For nanometer-sized particles, which can easily slip between the openings in the network of filter fibers, electrostatic attraction is the main way by which mask layers remove low mass particles, which are attracted to and bind to the fibers. This filtering of particles by electrostatic attraction is generally most efficient at low speed of the particles such as the speed of aerosols released by breathing through a face mask.
It is important to note that openings and gaps (such as those between the mask edge and the face) can compromise the performance. Findings indicate that leakages around the mask area can reduce efficiencies by ∼50% or more, pointing out the importance of a proper “fit”8.
Although a home-made fabric mask will at least offer some degree of protection against larger droplets and prevent access to facial features, it will not be very effective in protecting against respirable particles and droplets with a diameter of 0.3 to 2 μm, as these pass through the materials largely unfiltered5.
Thus, the inhalation of droplets containing viruses can be prevented by using a tight-fitting mask with particle filtering properties (self-protection). The FFP2/FFP3 mask type is very well suited to protect people from an infection by means of aerosol even when the environment is strongly contaminated with infectious droplets5.
How does mask structure affect filter particles?
For high filtration and blocking efficiency, the construction of masks layers is very important. Factors that contribute to this efficiency are these4,8:
Movement of droplets/aerosols is directly affected by interfiber spacing of the mask material and the number of layers. Combining layers of differing fiber arrangement to form hybrid masks uses mechanical filtering and may be an effective approach.
Electrostatic interaction impeding aerosol transmission is influenced by the type of mask material. Electrostatic attraction mainly affects the removal of low mass particles, which are attracted to and bind to the fibers. Leveraging electrostatic filtering may be another effective approach8.
The SEM pictures below show the structure and construction of mask fibers and give an insight into the factors that contribute to their high filtering and blocking efficiency.
An FFP2 mask combines layers featuring different spacing and fiber network types to form hybrid masks, employing both mechanical and electrostatic filtering.

Why are FFP masks superior?
Surgical and respiratory masks are compliant to regulations that guarantee to fulfill certain standards (cf. Table 1). The superior protection of FFP masks stems partially from its filtering layer (cf. Figure 3), using electrostatic filtration to block smaller particles (~0.1 µm).
Conclusion
While maintaining a safe distance from an infected or possibly infected person will prevent spread of infection through direct contact and droplet transmission, maintaining a safe distance may not effectively prevent the spread of infection through airborne aerosols. This is where it becomes very important to wear a mask.
Masks offer self-protection and minimize transmission of potentially infectious exhaled droplets to the surrounding atmosphere. However, in some situations like closed rooms or highly contaminated places, only masks with high blocking and filtration efficiencies will offer this kind of protection, provided they are closely fitted to prevent air from flowing around the mask edges.
The authors would like to explicitly thank Carl Zeiss GmbH, who provided the microscopic images.
References
1. Anand, S. & Mayya, Y. S. Size distribution of virus laden droplets from expiratory ejecta of infected subjects. Sci. Rep. 10, 1–9 (2020).
2. Chirizzi, D. et al. SARS-CoV-2 concentrations and virus-laden aerosol size distributions in outdoor air in north and south of Italy. Environ. Int. 146, 106255 (2021).
3. Lee, B. U. Minimum sizes of respiratory particles carrying SARS-CoV-2 and the possibility of aerosol generation. Int. J. Environ. Res. Public Health 17, 1–8 (2020).
4. Sanchez, A. L., Hubbard, J. A., Dellinger, J. G. & Servantes, B. L. Experimental study of electrostatic aerosol filtration at moderate filter face velocity. Aerosol Sci. Technol. 47, 606–615 (2013).
5. Kähler, C. J. & Hain, R. Fundamental protective mechanisms of face masks against droplet infections. J. Aerosol Sci. 148, (2020).
6. Oct, U. COVID-19 Scienti c Brief : SARS-CoV-2 and Potential Airborne Transmission small particles that can move through the air The term “ airborne transmission ” has a specialized meaning in public health practice respiratory microbes The epidemiology of SARS-Co. 2019–2022 (2021).
7. https://www.bfarm.de/SharedDocs/Risikoinformationen/Medizinprodukte/DE/schutzmasken.html Accessed 21 April 2021.
8. Konda, A. et al. Aerosol Filtration Efficiency of Common Fabrics Used in Respiratory Cloth Masks. ACS Nano 14, 6339–6347 (2020).
Für diesen Beitrag exisitiert leider keine deutsche Übersetzung.
Introduction
This protein is known under many different names such as non-structural protein NSP1, leader protein, host translation inhibitor and host shutoff factor. Some of these names already tell us about the function and importance of this relatively small protein. It is found in all betacoronaviruses1 and, even though it only contains 180 amino acids2, it is indispensable for the viral life cycle and the pathogenicity of SARS-CoV-2.
It plays an important role when it comes to the point where the virus needs its own genetic information in form of a string of codons. Its mRNA is translated into the corresponding amino acids that make up the viral proteins. Translation occurs either shortly after the virus entered the host cell (see life cycle) or after the viral mRNA has been replicated (as described here).
For this process, the virus does not have its own proteins; instead, it just uses the already existing translation machinery of the host cell: the ribosomes.
As ribosomes are responsible for synthesizing proteins by translating the information on the host’s mRNA into a string of amino acids, they are an important part of human cells. They consist of ribosomal RNA (rRNA) and ribosomal proteins, which form a larger (60S) and a smaller (40S) subunit3.
Here, the NSP1 comes into play. It helps the virus hijack ribosomes and use them for the replication of its own mRNA, while the host cells translation is supressed/inhibited/shut off4.
To understand how the NSP1 is involved in all this, we will first have take a closer look at the structure of the protein.
Structural features & interaction with ribosomes
Even though the full-length structure of NSP1 is unknown so far, we know what the two individual domains (connected via a linker that is 20 amino acids long) of the SARS-CoV-2 NSP1 look like and can even say a lot about its interaction with human ribosomes.
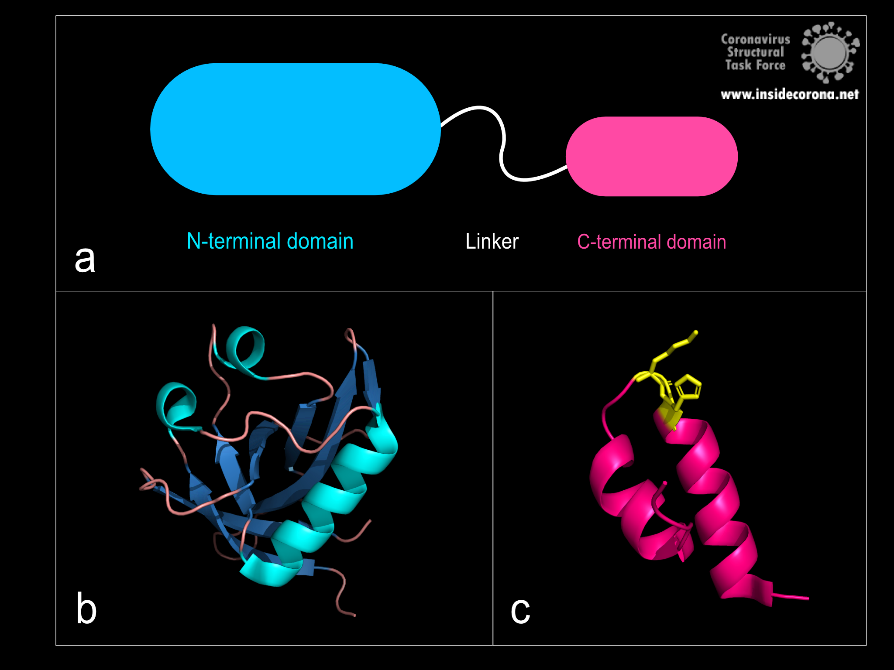
Figure 1: a: Schematic structure of NSP1. b: N-terminal domain (PDB: 7K7P)., c: C-terminal domain. KH motif (amino acids K164 and H165) in yellow (PDB: 6ZLW).
The first domain is the globular N-terminal domain (amino acids 1–128), which takes up most of the protein. It consists of a β-barrel of seven β-strands, two 310 helices and one α-helix5, as can be seen in Figure 1b.
The probably more interesting domain, due to the crucial role it plays for interaction with human ribosome, is the C-terminal domain comprising three moieties (Figure 1c). It consists of the two α-helices, α1 and α2, and a loop connecting them4. The shape of this C-terminal domain and its surface charge matches the mRNA entry channel of the ribosome perfectly and therefore covers the whole usual mRNA path4. In Figure 2, the small 40S ribosomal subunit (green) in a complex with the C-terminal domain of NSP1 (pink) is shown.
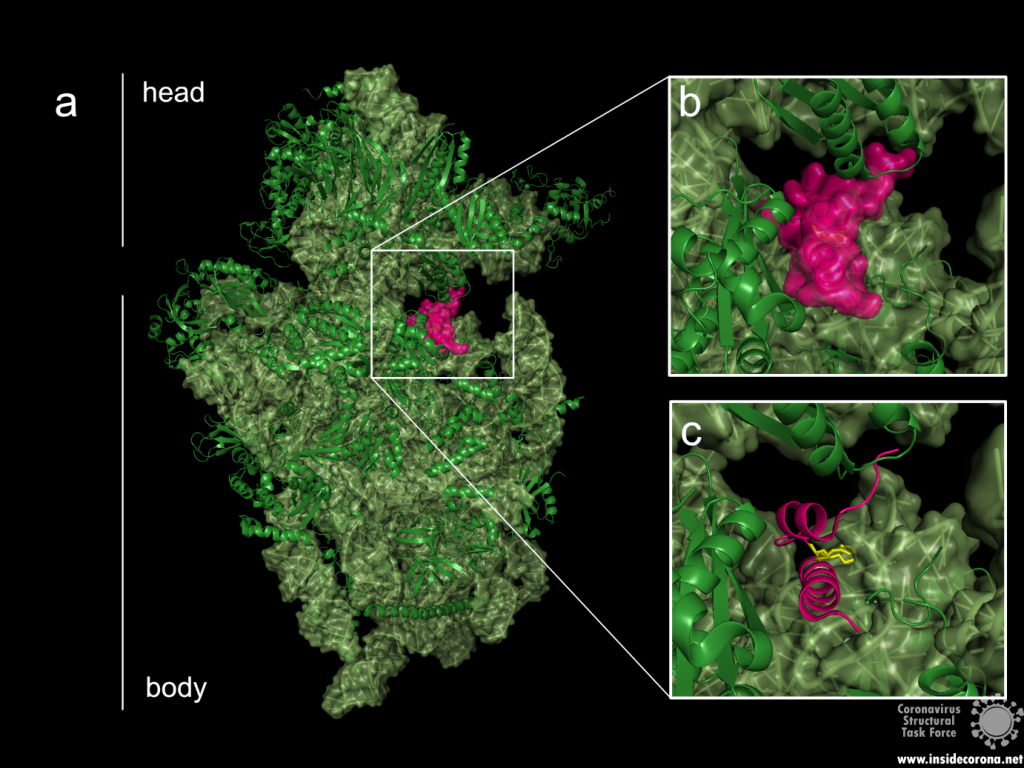
Figure 2: a: Ribosomal 40S subunit in complex with the NSP1 C-terminal domain (PDB: 6ZLW). The C-terminal domain is bound to the mRNA channel between the “head” and “body” of the 40S. b & c: NSP1 C-terminal domain shown with and without surface.
While the C-terminal domain is bound to the mRNA entry channel of the host cell’s 40S ribosomal subunit, the N-terminal domain can move around it within a 60 Å radius, connected by the 20 amino acid long flexible linker6.
All these interactions lead to an inhibition of the translation of the hosts mRNA—but how does the viral mRNA get translated, if the NSP1 is bound to the ribosome’s mRNA entry channel?
Viral translation
The virus needs a mechanism to circumvent its own translational blockage to maintain the capability for translation of the viral mRNA. It is not yet completely clear how this is accomplished, but different suggestions exist.
The first theory involves the N-terminal domain of NSP1 and the 5’ untranslated region (5’UTR) of the viral mRNA7.
In most coronaviruses, the 5’UTR part of the viral mRNA is conserved with a complex secondary structure6. Some scientists7 suggest that it might interact with the N-terminal domain, making the interaction between NSP1 and the ribosome sterically impossible and therefore lifting the blockage. This was also based on their study indicating that the C-terminal domain alone can suppress the host’s protein synthesis, but the N-terminal domain is needed to bypass the translation inhibition. Also, extending the linker between the two domains artificially by additional amino acids could be shown to reduce the viral mRNA translation7.
The second theory suggests that the translational blockage induced by the viral NSP1 is not lifted. In this mechanism, most ribosomes would be blocked by the NSP1s, but those left unblocked could still synthesize proteins. Here the viral 5’UTRs would make the mRNA of the virus more favourable than the host’s mRNA. This would lead the ribosomes into translating the viral mRNA with a higher efficiency than the cellular mRNA6.
Effect on the cells and immune system interference
Translation inhibition of the cellular mRNA by NSP1 results directly in another interesting and significant effect on the human cell. Besides the negative effects on normal cell functions, the translation of proteins involved in innate immune response is also inhibited. This includes interferons (proteins involved in antiviral activity8) like Interleukin-8, IFN-β, IFN-γ1 and anti-viral factors that are stimulated by interferons, leading to a downregulation of the cell’s defence system4,9.
Earlier studies on SARS-CoV-1 also showed that NSP1 is further inducing cleavage of the host’s mRNA, probably by using one of the host’s proteins. This again does not apply to its own viral mRNA10, making the impact on the host cell even greater.
Taken together, this protein is a major pathogenicity factor of SARS-CoV-2 and might therefore be an interesting drug target1.
Available structures
As of this writing, 16 structures of the SARS-CoV-2 NSP1 are available, of which two display the N-terminal domain. The other structures show the C-terminal domain in complex with a ribosome, ribosomal subunit or preinitiation ribosome. As there is no full-length structure solved so far, only predictions on the whole protein were made, for example given by Clark et al.5.
Available structures of the N-terminal: 7k7p, 7k3n.
Available structures of the C-terminal: 7k5i, 6zoj, 6zok, 6zm7, 6zlw, 6zmi, 6zp4, 6zon, 7jqb, 6zme, 6zmt, 6zn5, 6zmo, 7jpc.
References
- de Lima Menezes, G. & da Silva, R. A. Identification of potential drugs against SARS-CoV-2 non-structural protein 1 (nsp1). Journal of Biomolecular Structure and Dynamics 1–11 (2020) doi:10.1080/07391102.2020.1792992.
- Yoshimoto, F. K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. 19.
- Khatter, H., Myasnikov, A. G., Natchiar, S. K. & Klaholz, B. P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015).
- Thoms, M. et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. 8 (2020).
- Clark, L. K., Green, T. J. & Petit, C. M. Structure of Nonstructural Protein 1 from SARS-CoV-2. Journal of Virology 95, 12 (2021).
- Schubert, K. et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27, 959–966 (2020).
- Shi, M. et al. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. http://biorxiv.org/lookup/doi/10.1101/2020.09.18.302901 (2020) doi:10.1101/2020.09.18.302901.
- De Andrea M. et al. The interferon system: an overview. Eur J Paediatr Neurol (2002) doi:10.1053/ejpn.2002.0573.
- Vann, K. R. Inhibition of translation and immune responses by the virulence factor Nsp1 of SARS-CoV-2. 4.
- Huang, C. et al. SARS Coronavirus nsp1 Protein Induces Template-Dependent Endonucleolytic Cleavage of mRNAs: Viral mRNAs Are Resistant to nsp1-Induced RNA Cleavage. PLoS Pathog 7, e1002433 (2011).
Das Genom von SARS-CoV-2 codiert für ein langes Polyprotein ORF1a/ ORF1ab (ORF = offener Leserahmen). Es umfasst 16 nicht-strukturelle und vier strukturelle Proteine. Durch Leserasterverschiebungen (Engl: „Frame Shifts“) entstehen zusätzliche ORFs, die für sogenannte akzessorische Proteine codieren. Diese Gruppe von Proteinen steht im Verdacht einen großen Beitrag an der Pathogenese von SARS-CoV-2 zu leisten. Man nimmt an, dass eines dieser Proteine, das akzessorische Protein 7a, durch Induktion von apoptotischen Prozessen in menschlichen Wirtszellen maßgeblich an der Erkrankung Covid-19 beteiligt ist1.
Struktur von SARS-7a
Bislang ist es Forschern noch nicht gelungen die vollständige Proteinstruktur und Funktion von SARS-7a des neuen Coronavirus zu entschlüsseln. Das Protein besitzt aber in seiner Sequenz eine 85%ige Übereinstimmung und 95.2%ige Ähnlichkeit mit einem bereits bekannten Protein in SARS-CoV2. Somit kann angenommen werden, dass sich beide Proteine in Struktur sowie Funktion ähneln. Aus der Sequenzanalyse von SARS-CoV geht hervor, dass ORF7a für ein Typ I Transmembranprotein mit 122 Aminosäuren codiert3. Der N-Terminus umfasst ein Signalpeptid, bestehend aus sieben β-Strängen, die sich zu einem kompakten immunoglobulinartigen β-Sandwich, bestehend aus zwei β-Faltblättern, zusammenlagern (siehe Abbildung 1). Das erste β-Faltblatt umfasst die β-Stränge A, G, F, C und das zweite die Stränge B, E, D (Abbildung 1, links)4.
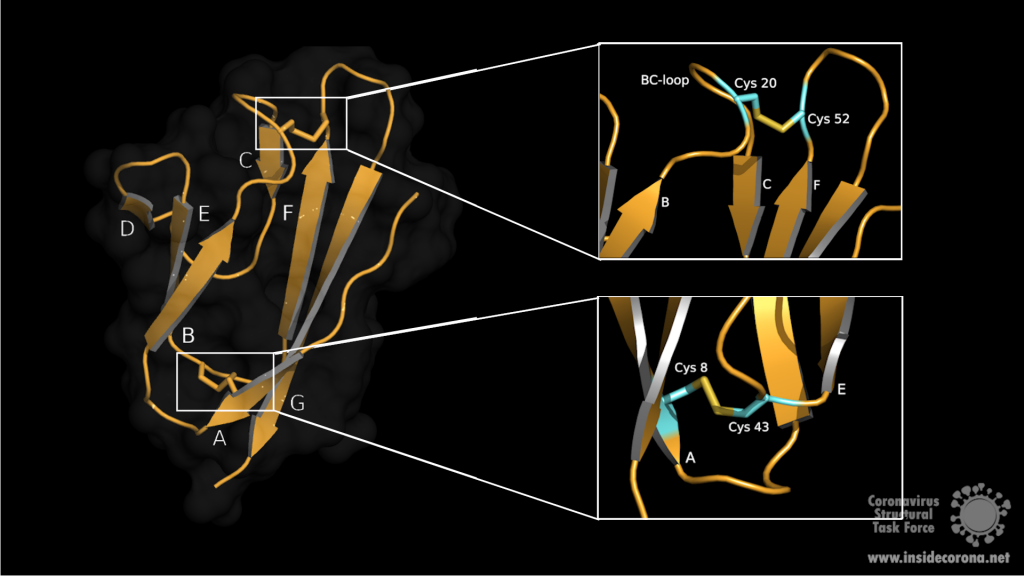
Die beiden amphipathischen Faltblätter sind eng aneinander gepackt, wobei die hydrophobe Seite innen liegt. Oberseitig wird die Ektodomaine über die β-Schleifen BC, DE und FG definiert. Die Unterseite bilden die Schleifen AB, CD und EF. Die Stabilisierung des β-Sandwichs erfolgt über zwei Disulfidbrücken. Die erste Disulfidbrücke liegt am unteren Ende der Faltblätter und verknüpft die Aminosäuren Cys8 auf Strang A mit Cys43 auf Strang E. An der Oberseite, auf der BC Schleife, liegt Cys20, welches über die zweite Disulfidbrücke mit dem Cys52 am Ende von Strang F verknüpft ist (siehe Abbildung 1, rechts). Oberhalb des BED-Faltblatts ragt die DE-Schleife aus dem β-Sandwich hervor und bildet mit den β-Strängen C und D eine Höhle. In dem Zentrum dieser hauptsächlich hydrophoben Vertiefung sitzt ein Glu18, das zur negativen Ladung am Boden der Höhle beiträgt. Aufgrund des negativen elektrostatischen Potentials ist es denkbar, dass diese Vertiefung eine potenzielle Interaktionsstelle für Liganden darstellt4.
Interaktionspartner von SARS-7a
Experimente mit Zellkulturen zeigen, dass SARS-7a vielfältige biologische Funktionen hat und auf unterschiedlichen Wegen in Zellprozesse eingreifen könnte5. Eine mögliche Schlüsselrolle ist die Zellzykluskontrolle. Die Überexpression von SARS-7a in HEK273 Zellen führte zu einer Inhibition des Zellwachstums und zum Zellzyklusarrest in G0/G1 Phase. Dieser Zellzyklusarrest kann die Virusreplikation begünstigen und die durch das Virus induzierte Pathogenität verstärken. Über die Interaktion von 7a mit einem Protein namens „B-cell lymphoma-extra large“ (Bcl-xL) kann Apoptose in menschlichen Nierenepithelzellen ausgelöst werden. Bcl-XL gehört zur B-cell lymphoma-2 Familie (Bcl-2), einer Gruppe von sogenannten „pro-survival“ Proteinen, die die Induktion der Apoptose inhibieren und so das Überleben der Zelle fördern. Durch die Wechselwirkung zwischen SARS-7a und der C-Terminalen Transmembrandomäne von Bcl-XL könnte dessen überlebenserhaltende Funktion unterdrückt und die Apoptose durch den Caspase-abhängigen Signalweg eingeleitet werden6,7.
SARS-7a kann auch eine Interaktion mit einer sogenannten Ap4a- Hydrolase, die in Prozesse der Zellproliferation, DNA-replikation, Apoptose und RNA-prozessierung involviert ist, eingehen und so deren Aktivität einschränken. Dieses Herunterregulieren der Hydrolaseaktivität führt zu einer gesteigerten Apa4 (Diadenosintetraphosphat) Produktion, wodurch es ebenfalls zu Apoptotischen Prozessen in der Zelle kommen kann5. Diese wirtszellenspezifische Regulation der Apoptose ermöglicht es dem Virus der Immunantwort zu entgehen und sich über weitere Organe auszubreiten.
Eine andere mögliche Funktion von ORF7a besteht in der Hemmung des Knochenmark-Matrix-Antigens 2 (BST-2), dass die Virusfreisetzung und so auch dessen Verbreitung, durch physisches Anheften der Virionen an die Plasmamembran, einschränken könnte. ORF7a antagonisiert diese einschränkende Wirkung durch die Bindung der extrazellulären Domäne von BST-2 wodurch dessen Glykosylierung verhindert wird. Wenn ein Wirkstoff gefunden wird, der in die ORF7a-BST-2 Interaktion eingreift, könnte dieser die Virusausbreitung verlangsamen oder sogar ganz stoppen8.
Zusammenfassung
Es lässt sich sagen, dass SARS-7a auf verschiedensten Wegen zur Pathogenität von SARS-CoV-2 beiträgt. Aus diesem Grund könnte die Entwicklung eines oder mehrerer Wirkstoffe, welche SARS-7a und dessen Interaktionen inhibieren, dabei helfen die Virusausbreitung zu verlangsamen und schwere Krankheitsverläufe verhindern.
6W37: Röntgenkristallstruktur des akzessorischen Proteins 7a, welches durch den Offenen Leserahmen ORF7a von SARSCoV-2 codiert wird.
1xak: Kristallstruktur des akzessorischen Proteins 7a von SARS-CoV. Das einzigartige typ I Transmembranprotein mit unbekannter Funktion hat eine kurze cytoplasmische Fraktion und eine Transmembrandomäne.
1y04: Röntgenkristallstruktur des akzessorischen Proteins X4, welches auch unter dem Namen 7a, U122 oder X4 bekannt ist. Das Protein lagert sich zu einem immunoglobulinartigen Betasandwich zusammen.
Weiterführende Literatur
- 1.Michel CJ, Mayer C, Poch O, Thompson JD. Characterization of accessory genes in coronavirus genomes. Virol J. Published online August 27, 2020. doi:10.1186/s12985-020-01402-1
- 2.Yoshimoto FK. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J. Published online May 23, 2020:198-216. doi:10.1007/s10930-020-09901-4
- 3.Fielding BC, Tan Y-J, Shuo S, et al. Characterization of a Unique Group-Specific Protein (U122) of the Severe Acute Respiratory Syndrome Coronavirus. JVI. Published online July 15, 2004:7311-7318. doi:10.1128/jvi.78.14.7311-7318.2004
- 4.Hänel K, Stangler T, Stoldt M, Willbold D. Solution structure of the X4 protein coded by the SARS related coronavirus reveals an immunoglobulin like fold and suggests a binding activity to integrin I domains. J Biomed Sci. Published online November 23, 2005:281-293. doi:10.1007/s11373-005-9043-9
- 5.Vasilenko N, Moshynskyy I, Zakhartchouk A. SARS coronavirus protein 7a interacts with human Ap4A-hydrolase. Virology Journal. Published online 2010:31. doi:10.1186/1743-422x-7-31
- 6.Tan Y-J, Fielding BC, Goh P-Y, et al. Overexpression of 7a, a Protein Specifically Encoded by the Severe Acute Respiratory Syndrome Coronavirus, Induces Apoptosis via a Caspase-Dependent Pathway. JVI. Published online December 15, 2004:14043-14047. doi:10.1128/jvi.78.24.14043-14047.2004
- 7.Tan Y-X, Tan THP, Lee MJ-R, et al. Induction of Apoptosis by the Severe Acute Respiratory Syndrome Coronavirus 7a Protein Is Dependent on Its Interaction with the Bcl-XL Protein. JVI. Published online April 11, 2007:6346-6355. doi:10.1128/jvi.00090-07
- 8.Taylor JK, Coleman CM, Postel S, et al. Severe Acute Respiratory Syndrome Coronavirus ORF7a Inhibits Bone Marrow Stromal Antigen 2 Virion Tethering through a Novel Mechanism of Glycosylation Interference. García-Sastre A, ed. J Virol. Published online September 16, 2015:11820-11833. doi:10.1128/jvi.02274-15
Leider steht für diesen Artikel keine deutsche Übersetzung zur Verfügung.
Introduction
It is known as VUI‑202012/01 or B.1.1.7 – the new mutation of the coronavirus Sars-CoV-2. It may be responsible for a sharply increased number of infections in the southeast of England (1), however, the scientific results leading to very strict lockdown measurements in the south of the UK, and travel restrictions across Europe are few and far between. Here, we have compiled what is known up until now.
On mutations
Mutations are normal in the evolution of life – and of viruses. If two similar viruses have infected the same cell, their genomes can become mixed-up, one of the reasons why animal influenza strains are considered so dangerous. This is also called recombination. Mutations can be caused by chemicals, radiation (including UV light) and errors during genome copying. A typical SARS-CoV-2 virus accumulates two amino acid changes per month in its genome — a rate of change about half that of influenza (2). This is because SARS-CoV-2 can repair RNA to some extent. But even so, this natural process led to thousands of mutations since the beginning of the pandemic. If they affected the virus life cycle negatively, that strain may have likely died out - if they did not make a difference or enhanced its chances of survival, it may have persisted.
Many mutations that are observed occur in the spike protein, which both serves to recognize potential host cells but is also what is being recognized by antibodies (i.e., the immune system).
Changes here can be crucial for the survival of the virus (“evolutionary pressure”) as they could significantly alter its affinity to the human receptor ACE2, which the virus uses as gateway to our cells.
What vaccines do
Most, if not all, potential COVID-19 vaccines expose our body to some part of the spike protein, which can be made by the body itself (mRNA vaccines) or carried by a harmless virus instead of SARS-CoV-2 (vector). Our body then produces antibodies which specifically recognize the spike and persist for several months. If we are exposed afterwards to the real virus, the body can recognize it immediately – and the risk of infection is much lower as the immune system swings into action immediately. Earlier this year, the spike mutation D614G (amino acid residue number 614 changing from aspartic acid (D) to glycine (G)) caused quite a stir in the media, and became the predominant form of SARS-CoV-2 (2, 3). However, if and in how far this was caused by natural selection is still debated (3). Another example which triggered an increased media coverage was the mutation Spike Y453F, which originated from infected minks in Denmark (4) and led to a culling of millions of animals. In any case, if we would be vaccinated with a spike protein form that would be different from the one in a virus we encounter later, there is a small chance that the vaccine may be rendered ineffective. This chance is, however, small for SARS-CoV-2, in any case much smaller than for HIV, which famously evaded any attempt to develop a vaccine.

What do we know?
There was a steep rise in infections in the UK recently, as in most other European countries.
A new mutation of the virus has emerged and seems to replace the old version of SARS-CoV-2 (5). Thousands of patients have been found to carry this variant.
This new variant has more mutations at once than expected. These mutations have not observed in this combination before.
The variant has been reported in the UK, the Netherlands, Denmark, Australia and Belgium so far.
What is striking to me as scientist about these findings is one thing in particular: How could the British government find that thousands of people were having the new SARS-CoV-2 variant, instead of the old, if the illness does not look any different? Sequencing samples from each and every patient would be technically very challenging, if not impossible. How could they know? The answer is:
Serendipity
The main PCR test employed in the United Kingdom is Thermo Fisher's TaqPathCOVID-19. This test identifies RNA on three different genome locations: In ORF1ab, nucleotide and spike. Now, it stopped working for the spike portion of the test, while the other two RNAs were still found to be present, which likely prompted scientists to sequence some of the samples in question. And indeed, the new mutant has a deletion of histidine-69 and valine-70, called 69-70del. This permitted easy differentiation of patients with the old SARS-CoV-2 (3 hits) and the new (2 hits) and is the reason why we know so much about the epidemiology of this variant!* It has also to be said that this test is not used as often in other countries, such as Germany, and this could well be the reason why we do not know if and how widespread it is here. In addition, other countries sequence much smaller proportions of virus isolates than the UK, so ongoing circulation of this variant outside of the UK cannot be excluded.
The details of the mutation
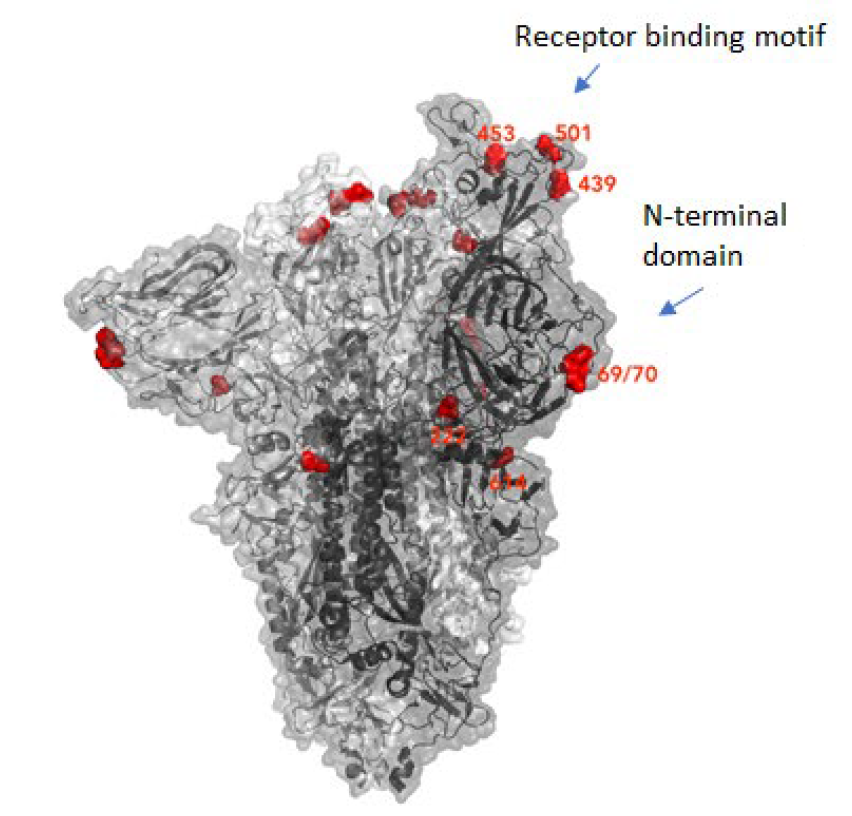
The new variant of SARS-CoV-2 VUI-202012/01 has 14 amino acid changes and three deletions affecting the genes for ORF1ab, spike and ORF8. One of these mutations (N501Y) occurs in the receptor binding domain and could lead to an increased binding affinity to the human ACE2. The 69-70 deletion has likely an immunological role and is the reason this mutant was detected so widely, as this RNA location is used for PCR tests. Another interesting mutation is the P681H, which is next to a furin cleavage site that has a biological significance in membrane fusion. These mutations could be responsible for the increased transmissibility. The effects of the other mutations aren’t fully investigated yet. Here is a list of the mutations which have been observed in the VUI‑202012/01 or B.1.1.7 variant:
| T1001I in gene ORF1ab | |
| A1708D in gene ORF1ab | |
| I2230T in gene ORF1ab | |
| SGF 3675-3677 deletion in gene ORF1ab | |
| A1708D in gene ORF1ab | |
| HV 69-70 deletion in spike | The 69-70 deletion on the spike protein is a re-occurring mutation that has shown to often co-occur with other amino acid changes in the RBD (6, 7). (1) Evasion to the human immune response and in association with other receptor binding domain changes (1) (2) Immunological role (8) (3) Leads to diagnostic failures which permit detection (see above, "Serendipity") (4) Associated with immune escape in immunocompromised patients (9(8)) Furthermore, the 69-70 deletion arose in multiple unrelated lineages and is associated with the evasion of the immune response (9). It is being hypothesized that this mutation undergoes a strong positive selection when exposed to convalescent plasma therapy in an immunocompromised human host (7). |
| Y144 deletion in spike | Deletion in the spike N-terminal domain (9) |
| N501Y in spike | One of six key contact residues in the spike receptor binding domains, this mutation leads to an increasing binding affinity to human and murine ACE2 (1). |
| A570D in spike | Mutation located at the spike receptor binding domain (10) |
| P681H in spike | The P681H mutation is located directly next to the furin cleavage site. It is one of the four residues which are insertions when compared to closely related coronaviruses, creating a furin cleavage site in the spike protein between the spike S1 and S2 domains. This prompts the entry of the virus into respiratory epithelial cells as well as the transmission in animal models (1) The S1/S2 furin cleavage site of SARS-CoV-2 is not found in closely related coronaviruses and has been shown to promote entry into respiratory epithelial cells and transmission in animal models (9) |
| T716I in spike | Mutation in in the S2 domain |
| S982A in spike | Mutation in in the S2 domain (10) |
| D1118H in spike | Mutation in in the S2 domain (8) |
| Q27 stop in ORF8 | The Q27stop mutation in the ORF8 leads to the truncation of the ORF8, and as it only consists of 121 amino acids, the consequence might be a loss of function. These and the other mutations could be responsible for the increased transmissibility of the B.1.1.7 variant. In any case, this mutation truncates the ORF8 protein at residue 27 or renders it inactive which allows further downstream mutations to accrue. (1) |
| R52I in ORF8 | |
| Y73C in ORF8 | |
| D3L in nucleocapsid | |
| S235F in nucleocapsid |
Why were there so many mutations at once?
This could be a result of prolonged or chronical SARS-CoV-2 infections as study of these infections reveal unusually large numbers of nucleotide changes and deletion mutations and often high ratios of non-synonymous changes. In addition to this, convalescent plasma treatment can cause intra-patient virus genetic diversity (11).
What does the new mutation mean in terms of impact and epidemiology?
There was an increase in cases with the new strain in total and in
proportion to the old (1). What does that mean for us?
This is what the internet says:
The COVID-19 genomics UK consortium (COG) reports about a “priority set of SARS-CoV-2 Spike mutations that are of particular interest based on potential epidemiological significance in the UK and/or biological evidence based on the literature or unpublished work.” (9)
The New and Emerging Respiratory Virus Threats Advisory Group of the British government (NERVTAG) discussed the new variant on Friday and concluded that its growth rate is higher by 67-75% and that this is likely due to a selective advantage. “In summary, NERVTAG has moderate confidence that VUI-202012/01 demonstrates a substantial increase in transmissibility compared to other variants.” (12) This is very likely the source of Boris Johnson’s claim to this strain being “70% more infectious”.
The English government writes that PHE (Public Health England) „is working with partners to investigate and plans to share its findings over the next 2 weeks. There is currently no evidence to suggest that the variant has any impact on disease severity, antibody response or vaccine efficacy. High numbers of cases of the variant virus have been observed in some areas where there is also a high incidence of COVID-19. It is not yet known whether the variant is responsible for these increased numbers of cases.” (13)
Conclusion
From this, we conclude that the British government, and we, do not know yet. It has not been conclusively shown that the new variant is more infectious (likely), has an easier time to evade the host immune system or if the vaccine will be less effective against it (very unlikely). The epidemologic model which predicts a higher tranmissability has still to be published, the science is still in the making. Tests of vaccines against the new variant are ongoing and will take a few weeks. There is yet little evidence that this new variant poses a significantly bigger threat than others - or to the contrary.
Acknowledgements
While I am listed as author of this article, it could not have been written without the help and research by Pairoh Seeliger, Lea von Soosten, Luise Kandler, Erik Nebelung and Oliver Kippes who all helped in this.
I would also thank Nicolai Wilk from Thermo Fisher Scientific who quickly responded to my questions about their test.
The title picture shows mutation cards from the game Pandemic Expansion: On the Brink by Z-Man Games.
- *The 69-70del mutation is predominantly observed in B.1.1 (including B.1.1.7), B.1.258, and the cluster 5 variant lineages of SARS-CoV-2.
References
- 1.A. Rambaut, Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. virological.org (2020), (available at https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563).
- 2.E. Callaway, The coronavirus is mutating — does it matter? Nature, 174–177 (2020).
- 3.L. Zhang, C. B. Jackson, H. Mou, A. Ojha, H. Peng, B. D. Quinlan, E. S. Rangarajan, A. Pan, A. Vanderheiden, M. S. Suthar, W. Li, T. Izard, C. Rader, M. Farzan, H. Choe, SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun (2020), doi:10.1038/s41467-020-19808-4.
- 4.ECDC, Detection of new SARS-CoV-2 variants related to mink. www.ecdc.europa.eu (2020), (available at https://www.ecdc.europa.eu/sites/default/files/documents/RRA-SARS-CoV-2-in-mink-12-nov-2020.pdf).
- 5.ONS UK , Percentage of COVID-19 cases that are positive for ORF1ab and N genes. www.ons.gov.uk (2020), (available at https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/adhocs/12690percentageofcovid19casesthatarepositivefororf1abandngenes).
- 6.R. M. Dawood, M. A. El-Meguid, G. M. Salum, K. El-Wakeel, M. Shemis, M. K. El Awady, Bioinformatics prediction of B and T cell epitopes within the spike and nucleocapsid proteins of SARS-CoV2. Journal of Infection and Public Health (2020), doi:10.1016/j.jiph.2020.12.006.
- 7.S. A. Kemp, D. A. Collier, R. Datir, S. Gayed, A. Jahun, M. Hosmillo, I. A. Ferreira, C. Rees-Spear, P. Mlcochova, I. U. Lumb, D. Roberts, A. Chandra, N. Temperton, K. Sharrocks, E. Blane, J. A. Briggs, K. G. Smith, J. R. Bradley, C. Smith, R. Goldstein, I. G. Goodfellow, A. Smielewska, J. P. Skittrall, T. Gouliouris, E. Gkrania-Klotsas, C. J. Illingworth, L. E. McCoy, R. K. Gupta, Neutralising antibodies drive Spike mediated SARS-CoV-2 evasion (2020), , doi:10.1101/2020.12.05.20241927.
- 8.K. Kupferschmidt, Mutant coronavirus in the United Kingdom sets off alarms, but its importance remains unclear. Science (2020), doi:10.1126/science.abg2626.
- 9.COG, COG-UK update on SARS-CoV-2 Spike mutations of special interest Report 1. https://www.cogconsortium.uk (2020), (available at https://www.cogconsortium.uk/wp-content/uploads/2020/12/Report-1_COG-UK_19-December-2020_SARS-CoV-2-Mutations.pdf).
- 10.S. Kemp, W. Harvey, R. Datir, D. Collier, I. Ferreira, A. Carabelii, D. L. Robertson, R. K. Gupta, Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion ΔH69/V70 (2020), , doi:10.1101/2020.12.14.422555.
- 11.ECDC, Threat Assessment Brief: Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. www.ecdc.europa.eu (2020), (available at https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-rapid-increase-sars-cov-2-variant-united-kingdom).
- 12.NERVTAG, NERVTAG meeting on SARS-CoV-2 variant under investigation VUI-202012/01. https://khub.net (2020), (available at https://khub.net/documents/135939561/338928724/SARS-CoV-2+variant+under+investigation%2C+meeting+minutes.pdf/962e866b-161f-2fd5-1030-32b6ab467896?t=1608470511452).
- 13.PHE, PHE investigating a novel variant of COVID-19 . www.gov.uk (2020), (available at https://www.gov.uk/government/news/phe-investigating-a-novel-variant-of-covid-19).
Für diesen Beitrag exisitiert leider keine deutsche Übersetzung.
This article has been written by Cameron Fyfe and Lea von Soosten.
In the previous two articles we spoke of proteins involved in RNA synthesis and proteins involved in removing errors during that process. There are also proteins produced by SARS-CoV-2 that can mimic functions of the host cell to avoid its defense mechanisms.
Eukaryotic cells have evolved to have various immune responses to fight infection or invasion from pathogens. One of these is to recognize and chop up any RNA that is from other organisms using enzymes called exoribonucleases. In order to differentiate "friendly" RNA from "foe" RNA is to give the cell's own RNA a VIP badge so that only unfriendly RNA will be shredded. These "VIP badges" are made of a 5’ to 5’ triphosphate linkage with two methylation modifications (see Fig. 1). In order to evade exoribonucleases, the virus SARS-CoV-2 has a way of 5’ to 5’ capping as well as adding its own methyl group VIP badges to protect its RNA from the defense mechanisms of invaded cells. Two Very Important Proteins, nsp14 and nsp16, have this methyltransferase activity using an S-Adenosyl methionine (SAM) as cofactor.
What are SAM methyltransferases?
Methyltransferase enzymes are a large superfamily of proteins that perform the chemical addition of a methyl group (a carbon with three hydrogens) to a variety of substrates. These substrates include small molecules, other proteins, DNA, and RNA 2,3. This superfamily of proteins often uses a small molecule, S-Adenosyl methionine (SAM), to transfer a methyl group to its target substrate (Figure 2). During this process, the methyl group bound to the charged sulfur is brought in proximity to the target atom of the substrate, transferring the methyl group (Figure 2), resulting in the methylated product and the byproduct S-Adenosyl homocysteine (SAH).
Methyltransferases of SARS-CoV-2
In a previous article we spoke of the exoribonuclease (ExoN) proofreading activity of Nsp14 (not to be confused with the host cell's own exoribonucleases that are part of the immune system, see above). After the 5’ to 5’ guanine triphosphate addition has been performed on the mRNA the guanine-N7-methyltransferase activity of Nsp14 comes into play producing the first Cap0 structure with a VIP tag (Figure 1, 3). Only after this methylation has been performed can Nsp16 have action and perform the second 2’O-methylation to produce the Cap1 structure (Figure 1, 3).
Not only do both of these proteins perform VIP methylations of mRNA, but they also both bind another non-structural protein, Nsp10. The binding of Nsp10 has been shown to increase activity in both Nsp14 ExoN activity and Nsp16 methyltransferase activity4. Independently, Nsp10 has also been shown to have the ability to bind both single and double stranded DNA and RNA5.
Structures of nsp14 and nsp16
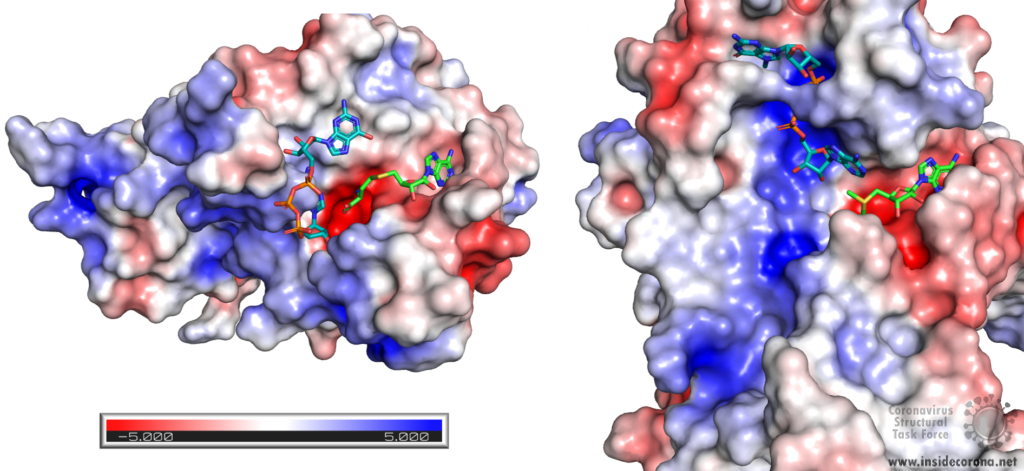
Nsp14 consists of two domains, each carrying out one specific task: the first is responsible for the ExoN activity, whilst the second executes the first methylation of the Guanosine-N7 of the RNA end cap. The two domains are connected by a flexible region that acts like a hinge, allowing movement between the domains. The second domain has an unusual and unique structure which does not follow the typical Rossmann fold seen in other SAM methyltransferases. The methyltransferase active site has a negatively charged binding pocket that holds SAM (SAH in Figure 4. A) in close proximity to the Guanosine-P3-adenosine-5',5'-triphosphate (GpppA) substrate (Figure 4A). The binding pocket holding the GpppA has a positive charge and the surface charge of the region below is also positively charged (Figure 4A). The distance between the N7 of the 5’ Guanosine and the sulfur that transfers the methyl group is 4.4 Å5,6. This close proximity of cofactor and substrate facilitates the methylation.
Similar to Nsp14, Nsp16 has a negatively charged binding pocket to position SAM in close proximity to the m7GpppA substrate (Figure 4. B). The m7GpppA binding site has a positive charge. The space nearby the 3’ end of the m7GpppA also has an overall positive charge and would be expected to bind the extension of the full length RNA (Figure 4. B)4. The distance between the methyl group and the sulfur of SAM and the 2’O of the m7GpppA substrate is 3.1Å and 4.9Å, respectively.
Structure of nsp10 and its function
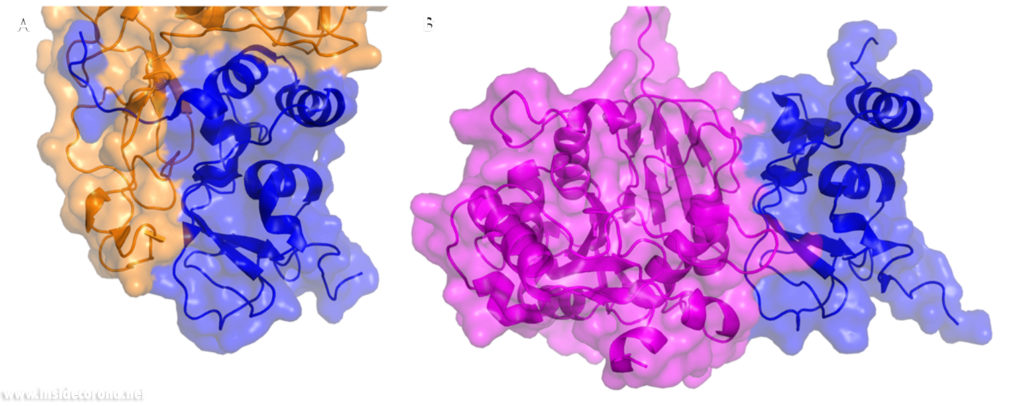
In a previous article where we spoke about the exoribonuclease (ExoN) activity of the first domain of nsp14, we highlighted the interaction between nsp14 and nsp10 (Figure 5A). This is quite significant, as the activity of ExoN increases 30-fold when nsp10 and nsp14 are bound. Nsp10 also functions as a co-factor for nsp16, stabilizing the SAM-binding pocket7 and enhances its methyltransferase enzymatic activity significantly4 (Figure 5B). For SARS-CoV, and similarly for MERS-CoV, the affinity for m7GpppA-RNA and m7GpppA cap analogue of nsp16 was found to be low until binding to nsp10, which enhanced the affinity for binding to RNA8,9. With a reduced activity in Nsp16 in the absence of Nsp10 and a huge decrease in activity of the exonuclease domain of Nsp14, interfering with these interactions could result in decreased viability of COVID-19.
Methyltransferases Nsp14 and Nsp16 as drugs targets
As both Nsp14 and Nsp16 use the cofactor SAM and have affinity for the endcap of RNA, these two binding sites could be worthwhile targets for drug development in the fight against SARS-CoV-2. Without the VIP status provided by the methylation of RNA the host immune system could defend against the viral RNA. It might be possible to block these binding pockets by letting the protein bind to something that is similar to SAM, which cannot function as a methyl donor. An additional challenge is that the inhibitor has to be very specific to Nsp14 or Nsp16, so as not to affect similar human proteins in a negative way.
Sinefungin is a 5’-aminoalkyl analog of SAH and SAM, which can do exactly that: it has the ability to inhibit all SAM methyltransferases (Figure 6). Sinefungin was first discovered in 1973 from Strepromyces griseolus and was described as having antifungal antibiotic properties10.
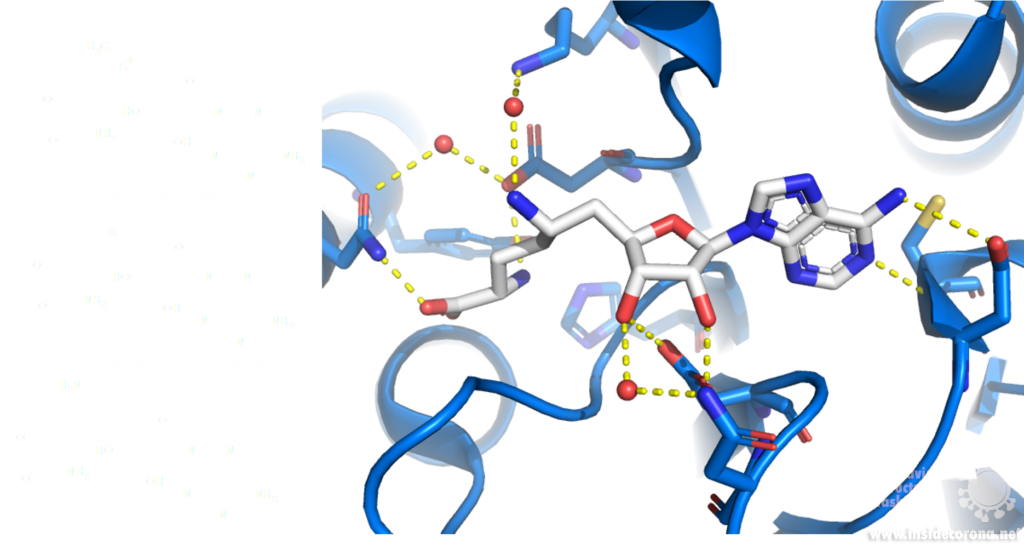
A major issue with targeting the SAM binding site of Nsps with compounds such as sinefungin (Figure 6) is that there are many proteins within humans that use SAM as a cofactor for normal function. This results in singefungin and other similar compounds having toxic effects on human cells. Synthetic chemists have already been able to synthesize analogs of sinefungin with improved affinities to specific SAM methyltransferases. Recently, specific inhibitors have been developed to target a nicotinamide SAM methyltransferase11. This inhibitor was developed to have affinity to both the cofactor binding site and the substrate binding site by combining the nicotinamide substrate with the SAM cofactor. Recent work has looked at how singefungin binds to the active site of Nsp16 in order to have a detailed understanding of its interaction to design more specific inhibitors that can target methyltransferases from SARS-CoV-24. Similar to the development of the nicotinamide SAM methyltransferase inhibitor, developing an inhibitor which binds to the substrate binding site as well as to the cofactor binding site could be effective. As Nsp14 and Nsp16 target different substrates, any inhibitors designed in this way would likely have specificity to only one of the two methyltransferases from SARS-CoV-2. Of the two, Nsp14 might be easier to target as it has a unique structure not similar to human SAM methyltransferases.
As both Nsp14 and Nsp16 interact with Nsp10 for normal function, interfering with this interaction could reduce activity of these enzymes. Further still, as the interface between Nsp10 with Nsp14 and Nsp16 has overlap the target is smaller for blocking binding of these proteins.
One way to look for possible drugs is repurposing those which are already approved for other diseases. Initial screen can be done in silico, by simulations of the interaction between the protein and the already existing and approved drug. However, such studies are highly dependent on the protein structures employed being correct, which is why we are evaluating all structures that are published for SARS-CoV and SARS-CoV-2.
Available structures
If you would like to look at the currently available structures for Nsp10, Nsp14, and Nsp16, they are available from our data base; we provide information on the quality of measurement data and models as well as improved structures.
All structures available for Nsp14 are bound to Nsp10 and are only available from SARS-CoV. The highest resolution structure of Nsp14 is PDB entry 5c8t at 3.2Å. It has a bound S-Adenosyl methionine ligand as well as zinc ions present. Alongside this, another structure of Nsp14 bound to S-Adenosyl homocysteine and a guanosine-triphosphate-adenosine ligand as well as zinc at 3.33Å resolution has been published (PDB: 5c8s). Additionally, two structures with zinc atoms but no ligands are available (PDB 5c8u 3.4Å at and 5nfy at 3.34Å). Both PDB entries 5c8t and 5nfy have been improved structures by our group.
Similar to Nsp14 all structures of Nsp16 are bound to Nsp10. There are currently 18 structures for Nsp16 bound to Nsp10 from SARS-CoV-2. The highest resolution structure is at 1.8Å and has SAM, Guanosine triphosphate and Adenosine bound as well as zinc atoms. The PDB:6wkq has Nsp16 bound to the methyltransferase inhibitor Sinefungin at 1.98Å resolution. Two further structures of note are 7jhe and 7jib that have various functional ligands. A further four structures are available from SARS-CoV.
Nsp10 alone: Currently there are two structures of Nsp10 from SARS-CoV-2, PDB 6zpe and 6zct, with the former having the highest resolution of 1.58 Å with bound zinc (PDB 6zpe). There are also three structures of Nsp10 from SARS-CoV available, PDB 2fyg, 2g9t, and 2ga6.
- 1.Ramanathan A, Robb GB, Chan S-H. mRNA capping: biological functions and applications. Nucleic Acids Res. Published online June 17, 2016:7511-7526. doi:10.1093/nar/gkw551
- 2.Boriack-Sjodin PA, Swinger KK. Protein Methyltransferases: A Distinct, Diverse, and Dynamic Family of Enzymes. Biochemistry. Published online December 22, 2015:1557-1569. doi:10.1021/acs.biochem.5b01129
- 3.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. Published online October 16, 2017:81-92. doi:10.1038/nrg.2017.80
- 4.Krafcikova P, Silhan J, Nencka R, Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun. Published online July 24, 2020. doi:10.1038/s41467-020-17495-9
- 5.Ferron F, Subissi L, Silveira De Morais AT, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci USA. Published online December 26, 2017:E162-E171. doi:10.1073/pnas.1718806115
- 6.Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc Natl Acad Sci USA. Published online July 9, 2015:9436-9441. doi:10.1073/pnas.1508686112
- 7.Rosas-Lemus M, Minasov G, Shuvalova L, et al. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. Published online April 20, 2020. doi:10.1101/2020.04.17.047498
- 8.Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells. Published online May 20, 2020:1267. doi:10.3390/cells9051267
- 9.Chen Y, Su C, Ke M, et al. Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex. Kuhn RJ, ed. PLoS Pathog. Published online October 13, 2011:e1002294. doi:10.1371/journal.ppat.1002294
- 10.Robert L. H, Marvin M. H. A9145, A NEW ADENINE-CONTAINING ANTIFUNGAL ANTIBIOTIC. J Antibiot. 1973;26(8):463-465. doi:10.7164/antibiotics.26.463
- 11.Policarpo RL, Decultot L, May E, et al. High-Affinity Alkynyl Bisubstrate Inhibitors of Nicotinamide N-Methyltransferase (NNMT). J Med Chem. Published online October 7, 2019:9837-9873. doi:10.1021/acs.jmedchem.9b01238
Für diesen Beitrag exisitiert leider keine deutsche Übersetzung.
COVID-19 is caused by the new coronavirus SARS-CoV-2. This virus has a characteristic virus hull featuring surface proteins which are commonly called “spikes”. Protruding from the viral hull like “spikes of a crown”, they give the coronavirus its name (corona = crown). These proteins make the first contact with human cells and are akin to keys that use a human receptor called “angiotensin-converting enzyme2” (ACE2) as a backdoor to gain access to and infect the cell.
1. Fuction of ACE2
ACE2 is a membrane protein which is anchored in the human cell membrane of epithelial cells. This type of cells can be found on the surface of lung, intestine, heart and kidney tissue. As a type I membrane protein, its primary function is to take part in maturation of angiotensin, a peptide hormone which controls vasoconstriction and blood pressure. ACE2 can be compared to a lock which can be unlocked by the coronavirus spike protein. The virus can then enter the cell and hijack its functions to reproduce itself, thus causing the Covid-19 infection which poses a serious danger to humanity, especially for older people and people with pre-existing conditions. For this reason, one approach to combating SARS-CoV-2 is to target and inhibit the spike to prevent infection. In order to do so, knowledge of the structural features of the spike and its interaction processes with ACE2 are indispensable. (Further information about how macromolecular structures are visualized can be found on our homepage: https://insidecorona.blogs.uni-hamburg.de/visualizing-macromolecular-structures/)
2. Spike: Structure and Fusion Mechanism
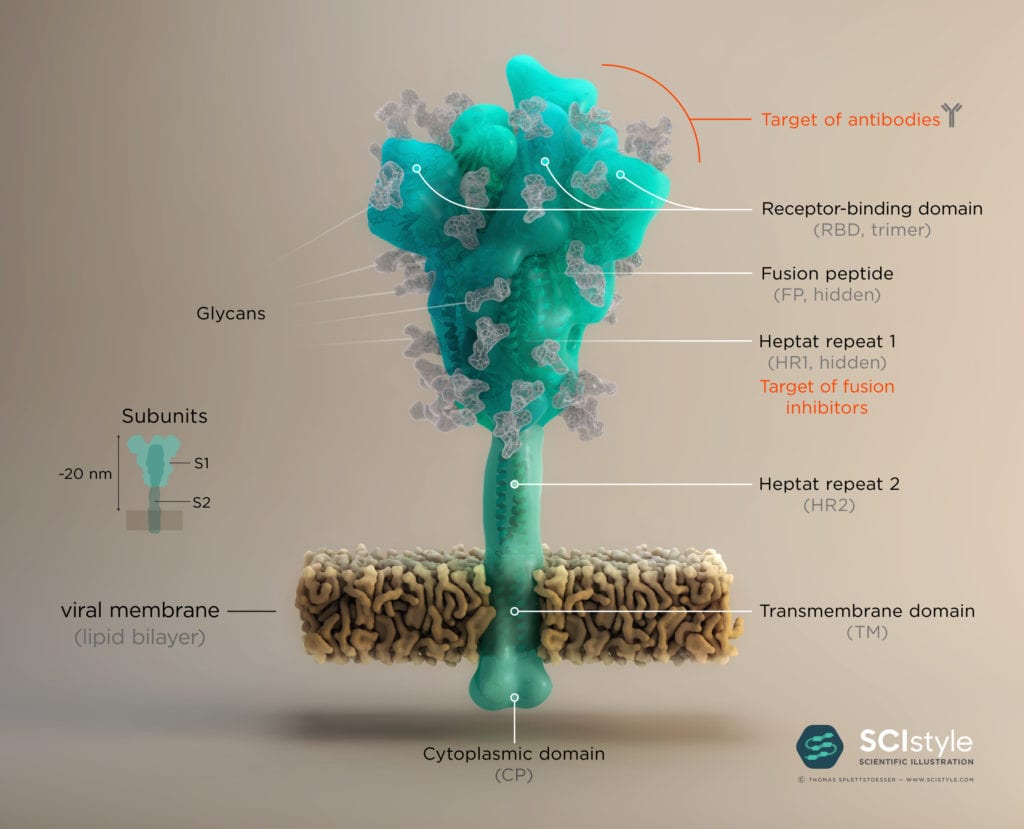
The Spike protein has a trimeric shape comprising three identical monomeric structural elements. Each of these monomers can fold out akin to a modern car key with a fold-out key element with specific teeth on its surface. This fold-out key element is the so-called “receptor binding domain” (RBD). The spike can only interact with ACE2 when its RBD is in a folded-out position, exposing its teeth, or “receptor binding motive” (RBM). As the name suggests, it comprises a motive of different amino acids which then can bind and unlock the ACE2 receptor. This key lock mechanism triggers a cascade of events initiating fusion with the host cell. First, protein scissors are recruited to the binding site. These scissors (furin & transmembrane serine protease 2) cleave the spike protein for subsequent activation. The active spike molecule then rearranges itself to form a long structural “hook” (formed of HR1/ HR2 and FP see Fig.2) that brings the epithelial cell and viral cell membrane into close proximity for fusion. Once the fusion is completed, the path for the virus is clear to transfer its genome encoded in ribonucleic acid (RNA) into the host cell. This successful transfer then enables the virus to multiply itself and finally spread from cell to cell, causeing Covid-19 in its wake.
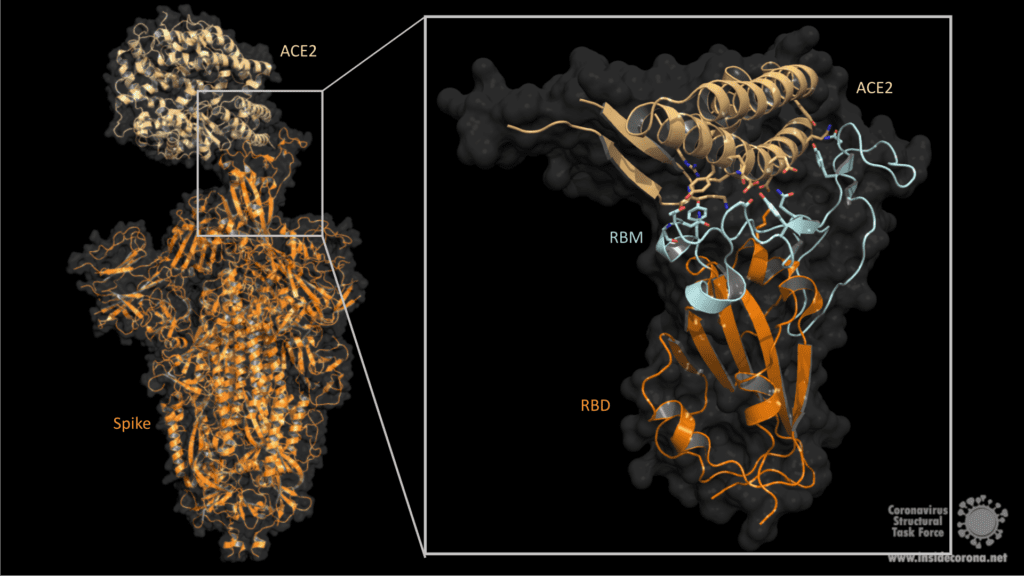
3. Evading the Immune System with Carbohydrate Chains
The human immune system normally recognizes the surface proteins of foreign organisms such as viruses or bacteria and reacts with an immune response to combat them. Spike proteins are such surface proteins but because of structural peculiarities, the coronavirus evades both the innate and the adaptive human immune system. The secret of these structural peculiarities are the N-glycans. These are long carbohydrate chains which sit on spike’s surface. Each spike comprises 66 N-glycans forming a protective shield around the protein. Hence the human immune system has problems recognizing spikes and identifying the coronavirus as an enemy.
The COVID 19 pandemic has a massive impact on our lives, our health and the global economy. Scientists around the world are trying to develop new drugs to combat the virus. Since the spike plays a critical role in the infection process, it is a prime target for drug development against the pandemic. One drug approach to inhibit the interaction between spike and the ACE2 receptor is to cap the spike protein using antibodies. Antibodies are proteins, normally produced by the human immune system to fight viruses. The idea is to treat patients with antibodies that cap the RBD of spike, thus preventing interactions with ACE2. This would lead to a nonfunctional spike, blocking the coronavirus from entering the cell (The key would no longer fit the lock). Another approach includes the development of small molecules that target and inactivate the protein scissor transmembrane serine protease 2 (see chapter 2), as the spike’s functionality depends on its cleavage activity. Since the spike protein decorates the virus hull, it could even be part of a potential vaccine. For this reason, the spike protein could also become the key in the molecular fight against COVID-19.
Für diesen Artikel exisitiert leider keine deutsche Übersetzung.
Crystallography has a problem. Some amino acid side chains in our structures simply can’t be seen in our maps (Fig. 1). Crystallographic maps represent many protein molecules in a crystal lattice, thousands of copies of the same molecule averaged over measurement time and unit cells. So, what happens with inherently flexible regions of our protein? The average of many different conformations leaves us with no map to guide us in modelling our side chain. So, what is the best way to deal with this as a model builder?
A passionate discussion within the Task Force has resulted in the following options for dealing with this situation:
- Set the occupancy of the unresolved atoms to 0
- Leave the atoms at full occupancy and allow the B-factors to inflate
- Trim the side chains to what can be resolved by the density
- Mutate the residue to a Proline, set your computer on fire, and walk away laughing maniacally.
Just to be clear, option four should only be considered in the direst of circumstances. Please consider options one to three before resorting to proline and fire, and even then, only with a computer you own. With that said, what is the best option? Sadly, none are ideal solutions to the problem so let’s discuss.
Option 1 can be misleading as the residue appears to be present in the model (Fig. 2), despite there being no experimental evidence for it, until you check the occupancy or load the corresponding map with your model which will tell you otherwise. An occupancy of zero also adds no useful information to the model and may even exclude atoms in this position, like opening the airlock and sending it flying out into the vacuum of space.
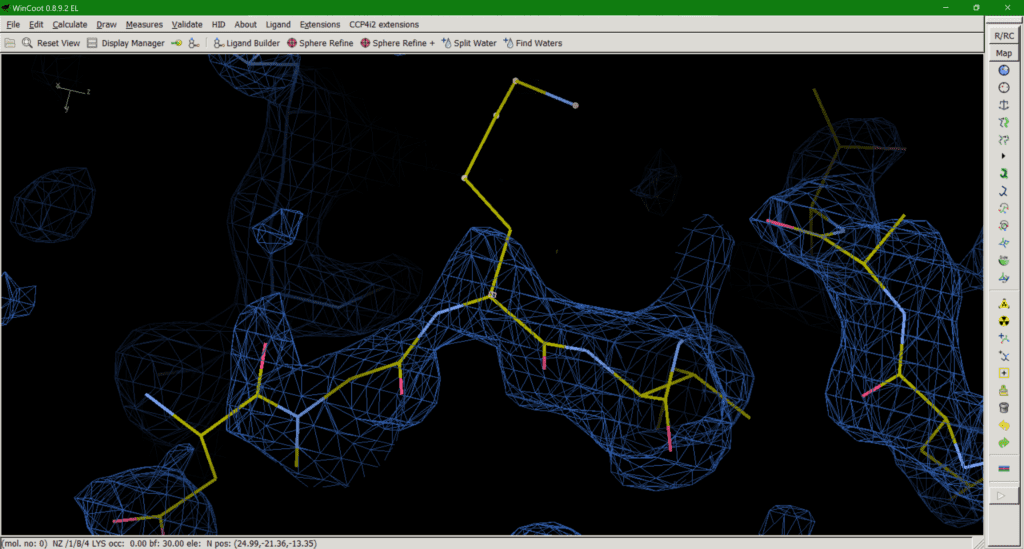
Option 2 is effectively the opposite of option 1, providing a full occupancy side chain in a sensible rotamer conformation and accept the resulting phase bias*. However, this can be equally misleading if the downstream user doesn’t check the B-factors of the sidechain, which will be very large, as they represent not only (smaller) displacement but (larger) disorder. In addition, allowing the B-factor to “explode” is not always an effective way to deal with this problem, as strong negative peaks can still be observed around the side chain in some cases. Another argument for maintaining an occupancy of 1 is that the protein sequence tells us a certain amino acid is present at a position, unless evidence of chemical clipping has been provided (mass spec, for example). Therefore, the atoms must be present in the protein so should be included in the model for the B-factors to deal with the physics of the situation. Options 1 and 2 both have the advantage of providing a complete set of atoms for downstream use in molecular modelling.
*During refinement our model will always bias the phase calculation which gives us our maps. Ideally, we would like out model to maximally affect the phases when we are confident our model is correct and minimally affect the phases when we are less confident. So, an occupancy of 1 (high confidence) where we observe no peaks in our map (low confidence) will lead to what we call phase bias. This can work both ways by underestimating the contribution of our model by setting the occupancy to 0 (option 1).
This brings us onto option 3: trimming down the side chain to what we can in the map (Fig. 3). The “make them work for it” option. If a downstream user is paying attention and realises that, for example, the side chain they are looking at is meant to be a lysine, despite the model only having atoms up to Cß, this should be the least misleading of all the options. The residue should not be mutated to, say, Alanine in this case, as that would mean you are wilfully misleading downstream users. Upon realising the atoms are missing, the downstream user can then model a (hopefully sensible) rotamer for their simulations if needed. The downside is that this approach does introduce some negative bias in favour of modelling bulk solvent into this area. Like I said, none of the options are ideal solutions.
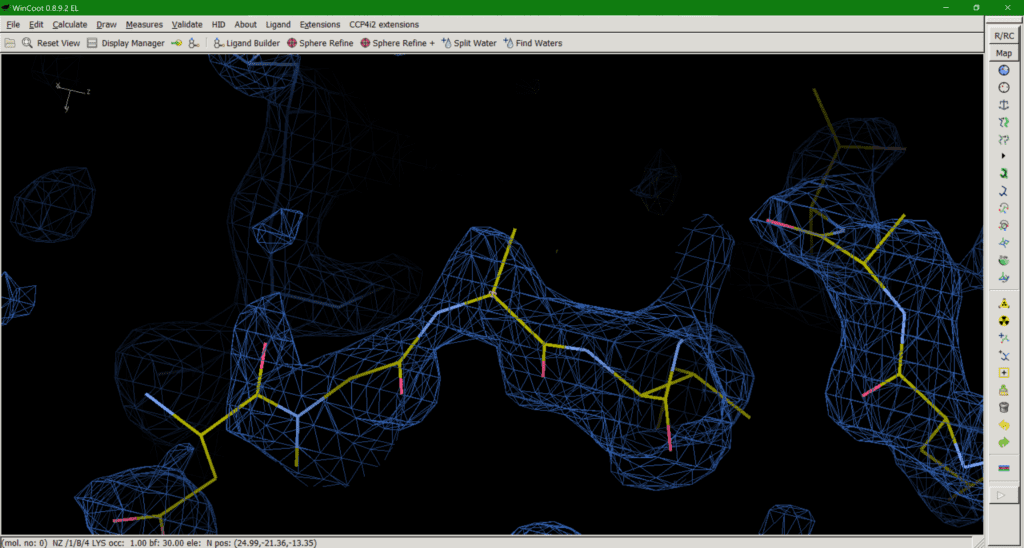
So, following this discussion between Nick Pearce, Dale Tronrud, Gianluca Santoni, Andrea Thorn, and I, we recommend option 3 as the best of the available solutions. We believe that the end goal of a crystallographic experiment should be to build atoms justified by the experimental data, i.e. the map, and leave the prediction of unobservable atoms to downstream users. We (crystallographers) are not here to “make it easier for users to avoid thinking about it”. However, after publishing the first iteration of this article a number of crystallographers made the case for option 2 on twitter and a poll of those involved resulted in 53.8% in favour of option 2 (Figure 4), so the matter is still far from resolved.
However, it’s nice to know that if we really can’t agree on the best method we can at least agree on not option 1, and there's always the fall back plan of option 4 and watch the PDB burn if we get desperate.

Für diesen Artikel steht leider keine deutsche Übersetzung zur Verfügung.
An important drug target
In the first part of this series we compared the protein nsp3 from SARS-CoV and SARS-CoV-2 by sequence. Now we delve deeper into the differences between these two proteins and follow through by analyzing the structure of one domain of nsp3 in particular: papain-like protease. This domain is a very relevant drug target because of its ability not only to cleave the polyprotein, but also remove some of the post-translational modification our cells use to fight these viruses. Without papain-like protease, the virus would be unable to spread COVID-19.
Like the entire nsp3 protein, the papain-like-protease (Pl2pro) domain is localized close to the endoplasmic reticulum’s (ER) membranes. The transmembrane domains hold it in place while the majority of the protein protrudes out of the ER membrane into the cytoplasm.[1]
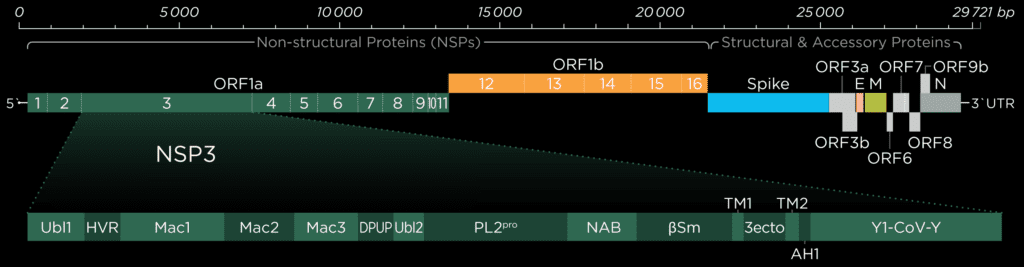
Ubiquitin-like-domain 2
We cannot discuss the Pl2pro domain without its little neighbor, which has been speculated to influence protease domain functionality.
In ubiquitin-specific proteases, the function of comparable Ubl2 domains is attributed to substrate recruitment or an increase in catalytic efficency. Ubiquitin-like-domain 2 (Ubl2) is the domain residing directly adjacent to the N-terminus of the Pl2pro catalytic domain. These ubiquitin-like domain seems to be more conserved compared to Ubl1 in different coronavirus species.[2]
If, in SARS-CoV and Murine coronavirus (MHV), Ubl2 is removed, Pl2pro loses its structural integrity. In addition, Pl2pro is then no longer able to act as an Interferon (IFN) antagonist (see below). However, some studies suggest that the Ubl2 domain in MERS-CoV might not be as essential as originally thought and in cell-based studies of this virus, Pl2pro could retain some of its enzymatic functions without the Ubl2 domain.[3]
To date, several inconsistent roles of Ubl2 were reported, and its exact function and inner workings remain enigmatic. This is being highlighted the fact that there are significant differences between the coronaviruses, and as a consequence, we need to exercise caution in applying our findings to SARS-CoV-2.
Combating the Host's Immune System
In the family of coronaviridae, viruses with either one or two Plpro domains can be found, with SARS-CoV and SARS-CoV-2 only having one. Confusingly, this single domain is however still called Pl2pro, even if it is the only papain-like protease domain in the viral genome.
Pl2pro cleaves the polyprotein from nsp1 (leader protein) up to nsp3. While Pl2pro cuts between nsp1-( ELNGG↓AV)-nsp2-( RLKGG↓AP)-nsp3-( SLKGG↓KI)-nsp4, the nsp5 (3c-like protease) cleaves the rest of the polyprotein. [2] The cysteine protease Plpro is similar to human ubiquitin-specific-protease (USP) in that it adopts a right-hand fold with "thumb", "palm" and "finger" subdomains.
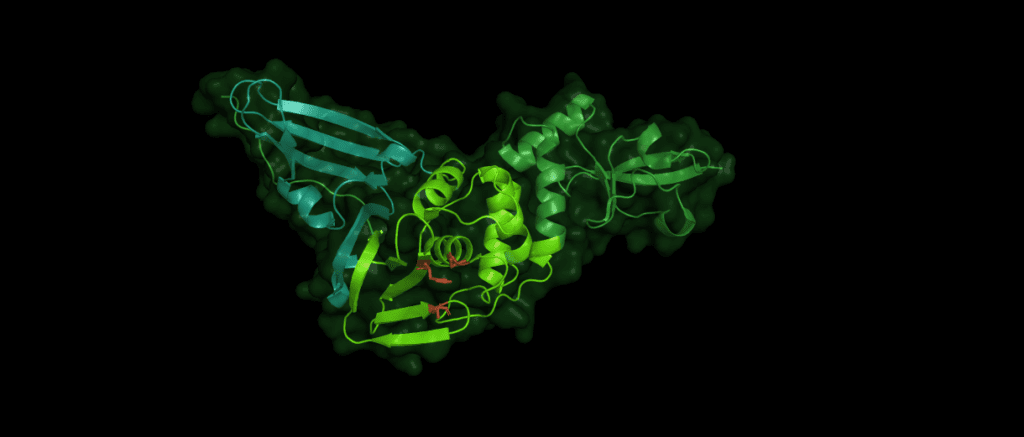
Despite the variations of Pl2pro in different coronaviridae, the same catalytic motif of three amino acid residues is essential for the stability and proteolytic activity of the domain: Cys112 is located in the thumb, His273 and Asp287 are located in the palm subdomain. (The numbers identifying these residues can vary between species.)
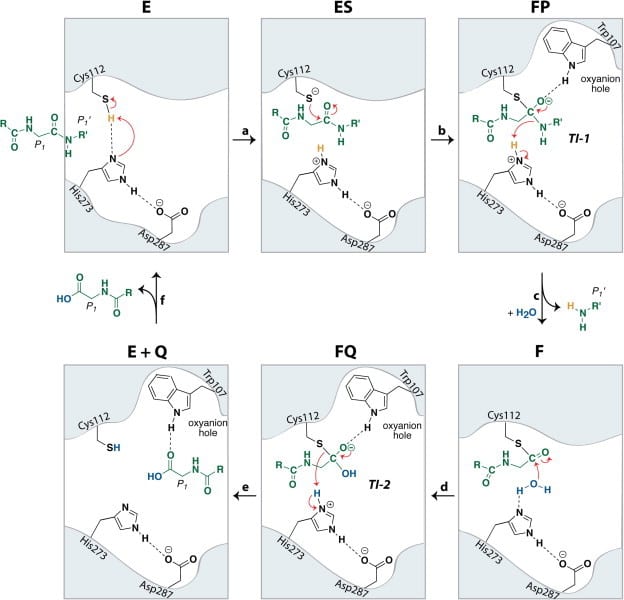
In addition, Pl2pro has deubiquitinating and deISGylating (removal of ISG15 from target proteins) abilities.[4] Both ubiquitin and ISG15 regulate facets of the immune response and through their removal Pl2pro poses as an antagonist to the human immune response. They can stimulate the production of cytokines, chemokines and other IFN-stimulated gene products which have antiviral properties. [6] ISG 15 is an ubiquitin-like modifier composed of two ubiquitin-like folds that has an essential role in marking newly synthesized proteins during the antiviral response.[3] Post-translational modification by ubiquitin and interferon-stimulating gene 15 (ISG15) is reversed by isopeptide bond hydrolysis. Figure 3 shows a proposed mechanism for the cleaving of isopeptide bonds by SARS-CoV.
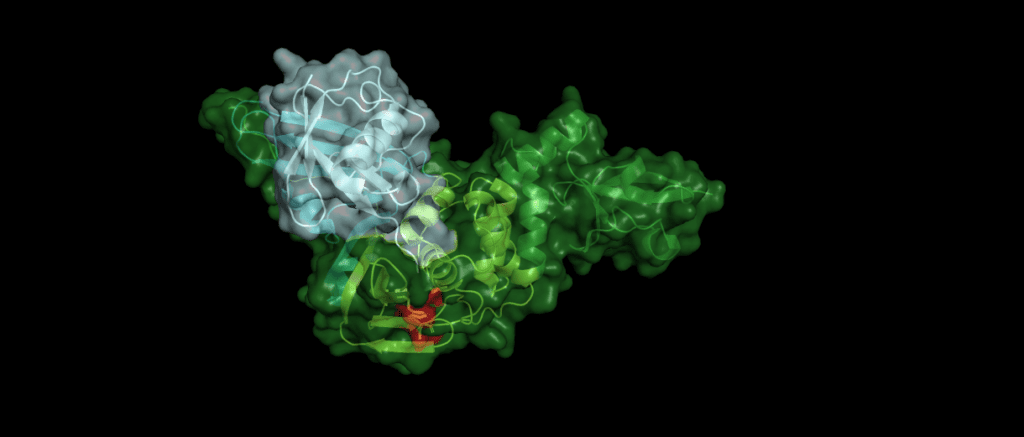
An example
Toll-like receptors (TLRs) are an important part of the machinery of the human immune response, which recognizes the pathogen-associated molecular patterns. The ability of the host cell to transduce the so-called Toll-like receptor 7 (TLR7) mediated immune response is diminished (Fig. 5) by Pl2pro as it removes Lys63-linked-ubiquitin from the TNF receptor associated factors TRAF3 and TRAF6. [5]
In addition, SARS-CoV can hamper the antiviral activities of interferon. The Pl2pro domain inhibits in combination with a transmembrane (TM) domain the STING mediated activation of interferon expression. PL2pro-TM interacts with TRAF3, TBK1, IKKε, STING and IRF3, the key components assembling a regulatory complex for activation of IFN expression.[5]
Another tool to fight the coronavirus in human cells is the "guardian of the genome", p53. The tumor supressor protein p53 impedes the replication of SARS-CoV, though the virus fights back with Pl2pro, which binds a p53 degradation stimulator named "RING finger and CHY zinc finger domain-containing protein 1" (or short: RCHY1). Enhanced by the Macro somains in NSP3, this binding enhances the stability of RCHY1 and hence promotes the degradation of p53. In addition, Pl2pro blocks another crucial cellular defense mechanism: The NF-κB pathway, which regulates immune responses to infections. SARS-CoV Pl2pro can stabilize IκBα, an inhibitor of NF-κB.[3]
Although all Pl2pro in different coronaviridae suppress the immune response, the targets differ between various species. For example, SARS-CoV Pl2pro preferentially processes Lys48 linked poly-ubiquitin chains, which are markers for proteasome degradation. MERS, on the other hand, shows no differences in effectivity between Lys48 and Lys63 linked di-Ubq chains. Lys63-linked chains are related to signal transduction cascades of the host immune system. Studies have shown that specificity among Pl2pro for Ubiquitin and ISG15 substrates can be altered with as little as a single amino acid change.[6] However, even though there are differences, for SARS-CoV-2, it is likely that at least some of the functions are similar.
Structural comparison
In order to predict Pl2Pro function for the novel Coronavirus SARS-CoV-2, we start by aligning their sequence like we did in the first part of this series to comapare the sequence with the one from SARS-CoV-2. Both domains share a similarity of 82.8% over the length of 313 amino acids. However, this time, we go for a more detailed analysis of the 54 individual differences, which are:
T3R N14I V20V N48N H49S V56Y D60N E66V D75T S77P P95Y G99N S114A V115T L116A L119T E123I K125L P129P A134D A143E N155C H170S L171Y Q173F S179D K181C C191T T195Q T196Q G200K N214E L215Q K216F G218K I221Q C225T D228K A229Q Y232K F240P Y250Q L252E Q254K G255H C259T E262S H274K K278S I284C L289L S293S T300I S308N
(The first letter refers to SARS-CoV, and the second to the amino acid residue in SARS-CoV-2.)
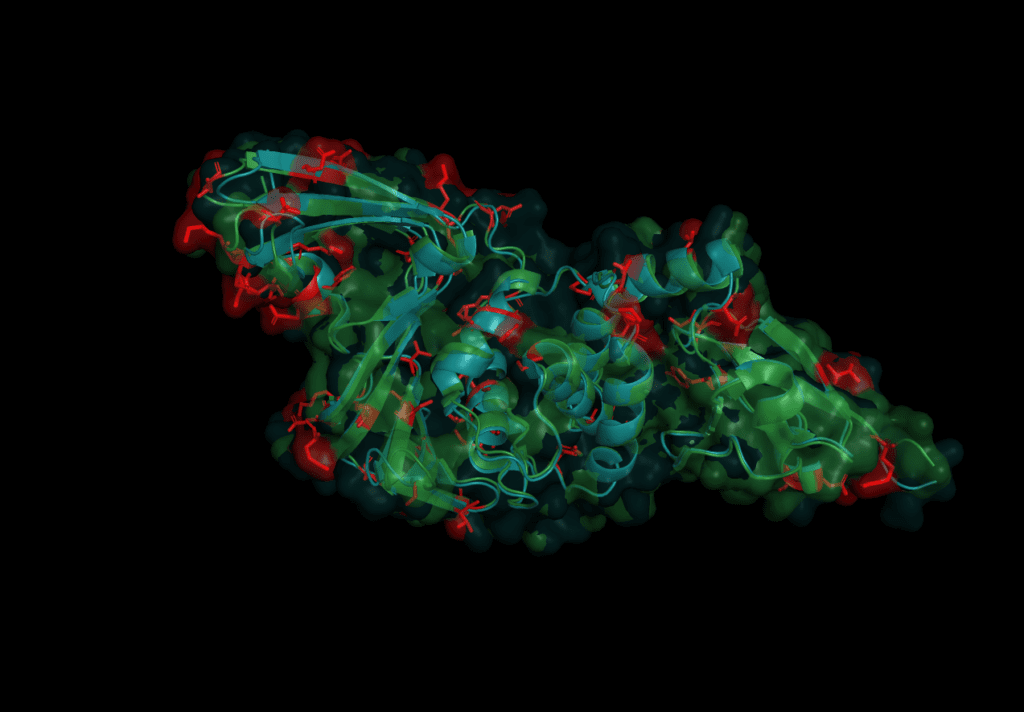
The mutations are evenly spread over the protein. None of the catalytic triad (Cys 112, His 273, Asp287) are changed as is to be expected given their conservation in all other coronaviruses. On further investigation, however, in the motif which interacts with ubiquitin six sites are different: S170T, Y171H, F216L, Q195K, T225V, and K232Q. Earlier studies concluded that the mutation of position 232 from Glutamine to Lysine increases the affinity for ubiquitin at the expense of the de-ubiquitination effectiveness.[6] The kinetics of SARS-CoV-2 nsp3 Pl2pro were studied to test if the protease domain of nsp3 has a reduced effectiveness in binding ubiquitin compared to nsp3 from SARS-CoV, MERS-CoV.
All three Pl2Pro variants cleave more ISG15 than ubiquitin. SARS-CoV has the fastest kinetics of the three viruses. And, the slower kinetics of SARS-CoV-2 resemble those of MERS-CoV rather more than SARS-CoV, having a 10 times higher turnover rate (kcat) as a deISGylase than as a deubiquitase.[6]
Besides the kinetics, the Pl2pro’s affinity for different poly-ubiquitin linkage sites was measured. The result shows that while SARS-CoV-2 can cut K48-Ub linked polyproteins, it seems to lack an ability to cut other polyubiquitin chains. Those K48-Ub linked polyproteins are cleaved at a slower rate than by SARS-CoV. In this regard, SARS-CoV-2 distinguishes itself from MERS-CoV which has the ability to cleave K63-linkages. It is suggested that the decrease in deubiquitinase effectiveness may not be irrelevant, but could lead to the often-mild symptoms that are a factor in why SARS-CoV-2 has been able to evade our efforts in quarantine. But this is mere speculation and a lot more research is needed to resolve the matter.[6]
PL2pro as a drug target
Pl2pro was a potential drug target early on in SARS-CoV-2 research. Hilgenfeld et al. name two major challenges we have to overcome to find a drug targeting Pl2pro. One is that the binding sites are tailor-made to bind glycine residues. Also, this very specific binding motif is rather ubiquitious in our cells. These two problems make it difficult to find an inhibitor which fits and is specific to Pl2pro. However, scientists found a weak spot: a loop called Blocking Loop 2 (BL2) regulates substrate binding and may be a promising target to inhibit PL2pro.[2] Naphthalene based inhibitors, which were earlier proposed to inhibit the BL2 of SARS-CoV, were shown to also inhibit SARS-CoV-2 Pl2pro, in particular an inhibitor called GRL-0617.[6]
For in-silico drug development, it might be prudent to choose high-resolution structures which already have a ligand or inhibitor bound, such as 6yva, 6wuu, 6wx4 or 6yaa. Technically speaking, 6wrh, albeit being a mutant, is one of the highest-quality structures available for SARS-CoV-2 Pl2pro.
In fact, a lot of research is still required to consolidate our understanding of this protein and its domains. In spite of that, we are making progress in our endeavor to fight this virus - and every step we take is one more to win this fight.
Sources
[1] Báez-Santos YM, St John SE, Mesecar AD. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21-38. doi:10.1016/j.antiviral.2014.12.015, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5896749/
[2] Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58-74. doi:10.1016/j.antiviral.2017.11.001, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7113668/
[3] Clasman JR, Báez-Santos YM, Mettelman RC, O'Brien A, Baker SC, Mesecar AD. X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility for structure-based drug design. Sci Rep. 2017;7:40292. Published 2017 Jan 12. doi:10.1038/srep40292, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5228125/
[4] Lei J, Hilgenfeld R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 2017;591(20):3190-3210. doi:10.1002/1873-3468.12827, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7163997/
[5] Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5(5):369-381. doi:10.1007/s13238-014-0026-3, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3996160/
[6] Freitas BT, Durie IA, Murray J, et al. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease [published online ahead of print, 2020 Jun 4]. ACS Infect Dis. 2020;acsinfecdis.0c00168. doi:10.1021/acsinfecdis.0c00168, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7274171/
Für diesen Beitrag exisitiert leider keine deutsche Übersetzung.
The world holds its breath as the novel Coronavirus continues to spread across the world, bringing our lives to a halt. We have gathered a lot of knowledge about the virus but there are still many gaps to fill. The non-structural-protein 3 (nsp3) represents one of these gaps in our knowledge. As the largest protein encoded by the coronaviruses genome, untangling its structure and function poses a huge task.
However, we can glean some knowledge around the specific function of SARS-CoV-2 nsp3 by looking at the virus‘s subfamily, Orthocoronaviridae. As related viruses do share some common traits, academics were not completely unprepared when SARS-CoV-2 came. In the background, while only very few people were worried about a new corona virus, scientists around the world had been investigating the invisible enemy for decades. Building on this past work we look at the functions of proteins from other coronaviruse, like Murine Hepatitis Virus (MHV) and SARS-CoV, to learn more about how best to fight against SARS-CoV-2.
The gene which produces nsp3 lies on the open reading frame 1a (ORF1a) which encodes polyprotein 1a. The sequence for nsp3 of SARS-CoV is 1922 amino acids long and sandwiched between nsp2 and nsp4. It not only cleaves itself from the polyprotein by its papain-like protease domain but also nsp1 and nsp2. In coronaviruses, 18 different domains have been found in nsp3. Each virus type has 10 to 16 of these, out of which eight domains and two transmembrane regions form the conserved part of nsp3, which can be found in every coronavirus known to date [1]:
- Ubiquitin-like-domian (Ubl1)
- Ubiquitin-like-domain (Ubl2)
- Papain-like protease (PlPro)
- Macro domain / X domain (Mac)
- Hypervariable region / Glu-rich acidic domain (HVR)
- Transmembrane regions (TM1)
- Transmembrane regions (TM2)
- Ectodomain / Zinc finger domain (3ecto)
- Nidovirus-conserved domain of unknown function (Y1)
- Coronvirus specific carboxyl-terminal domain (CoV-Y)
To start our investigation on SARS-CoV-2 related structural data, we will look into the protein sequences of SARS-CoV and SARS-CoV-2 to learn where they are similar and where they differ.
Genetic Comparsion of SARS-CoV and SARS-CoV-2
SARS-CoV has 16 domains which span 1922 amino acids. The nsp3 protein of SARS-CoV-2 is a bit longer at 1945 amino acids. When compared to each other, there is an overall similarity of 75,97%.[2] In Addition to the ten conserved domains the nsp3 gene of SARS-CoV-2 codes for four domains:

- Nucleic-acidic-binding domain (NAB)
- Betacoronavirus specific marker domain (βSM)
- Domain preceding Ubl2 and PL2pro (DPUP)
- Amphipathic helix 1 (AH1)
The two domains at the N-terminal end, Ubl1 and HVR, have an alignment of 79% and 64%, respectively. There seems to be a trend in coronaviridae for these domains to be poorly conserved, but Ubl1 still adopts the expected conserved fold.[4] If this proves true, could be analysed by comparing the sequence alignment and the structural similarity. It is unsurprising that the "high variable region" lives up to its name and shows the worst alignment of all. In the related MHV nsp3, this domain is dispensable for replication.[5]
It has been speculated that the Mac1 domain functions as an ADP ribose 1"-phosphatase, however, the effects of mutation in this region differ from virus to virus.[4] As a result, it is difficult to judge what significance the bad alignment of this domain will have on our understanding of SARS-CoV-2 without further research.
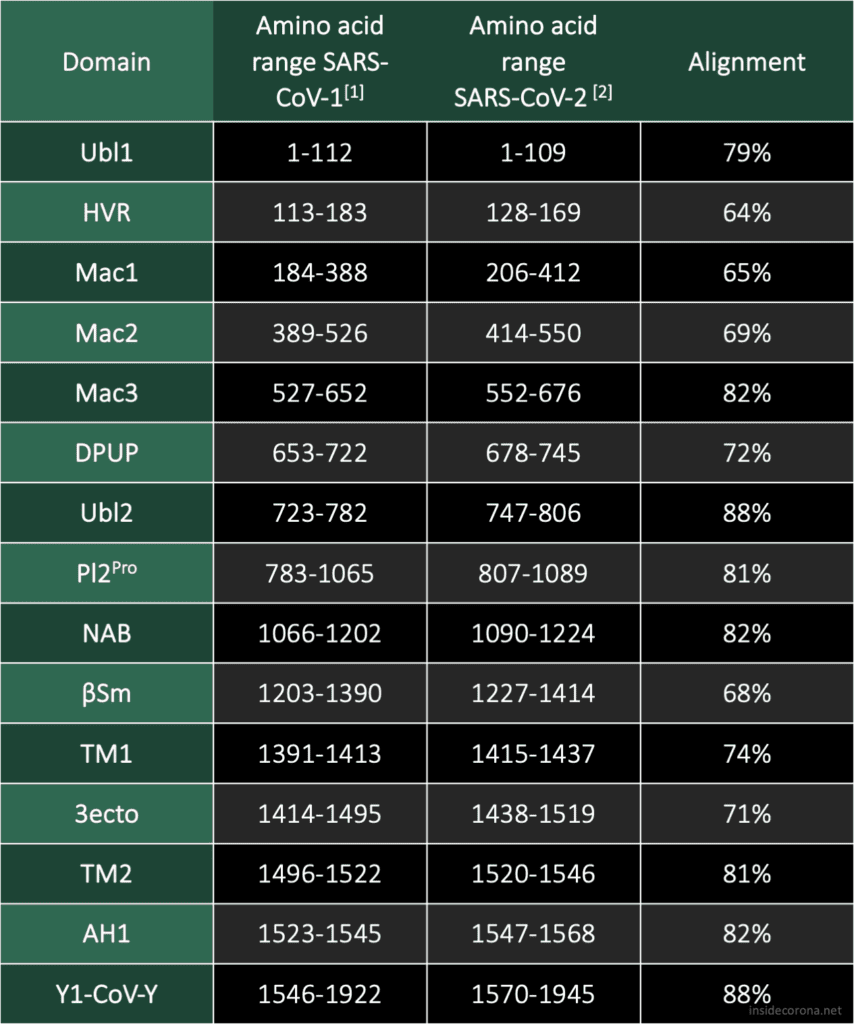
The Mac1 domain, also known as the X-domain, is followed by two macrodomains which were originally called "SARS-CoV Unique domains" (SUD-N and SUD-M), but were renamed when they were found to not be unique to SARS-CoV. It has since been observed that only Mac3 plays an essential role in viral RNA replication[6], which could explain why Mac3 is one the most conserved domains in the alignment of SARS-CoV and SARS-CoV-2.
Pl2Pro and its neighbouring domain Ubl2 show some of the highest sequence alignments of all domain comaprisons. This could be explained by their essential function to cleave nsp3 from the polyprotein.
Little is known about the domains following Pl2Pro and our current structural knowledge is limited to a nuclear magnetic resonance (NMR) structure of NAB. While the structure and function of Y1 and CoV-Y from SARS-CoV-2 are currently unknown, their sequence, which compromises a fifth of the genome, is highly conserved in all coronaviruses.
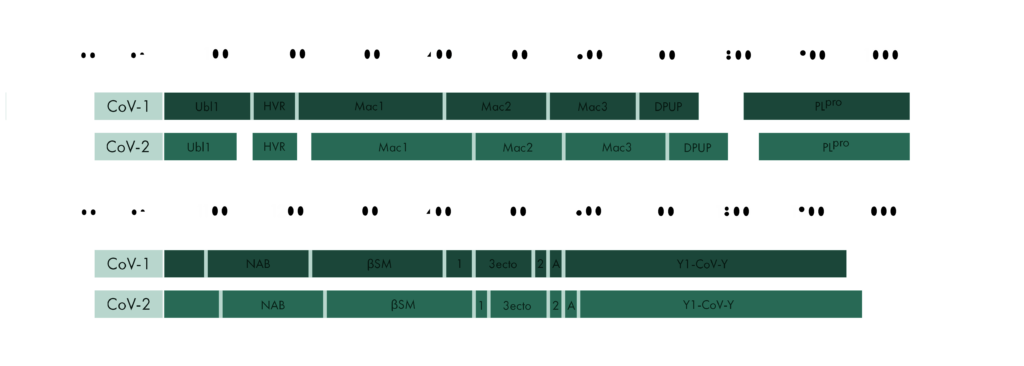
In the second part of the series of Untangling Nsp3 of SARS-CoV-2 we will delve deeper into some structures of nsp3 of SARS-CoV-1 and SARS-CoV-2 and will try to find out how the differences in the sequence may have influenced some structures of the protein. For a further in-depth reading on the topics discussed here I highly recommend the sources below.
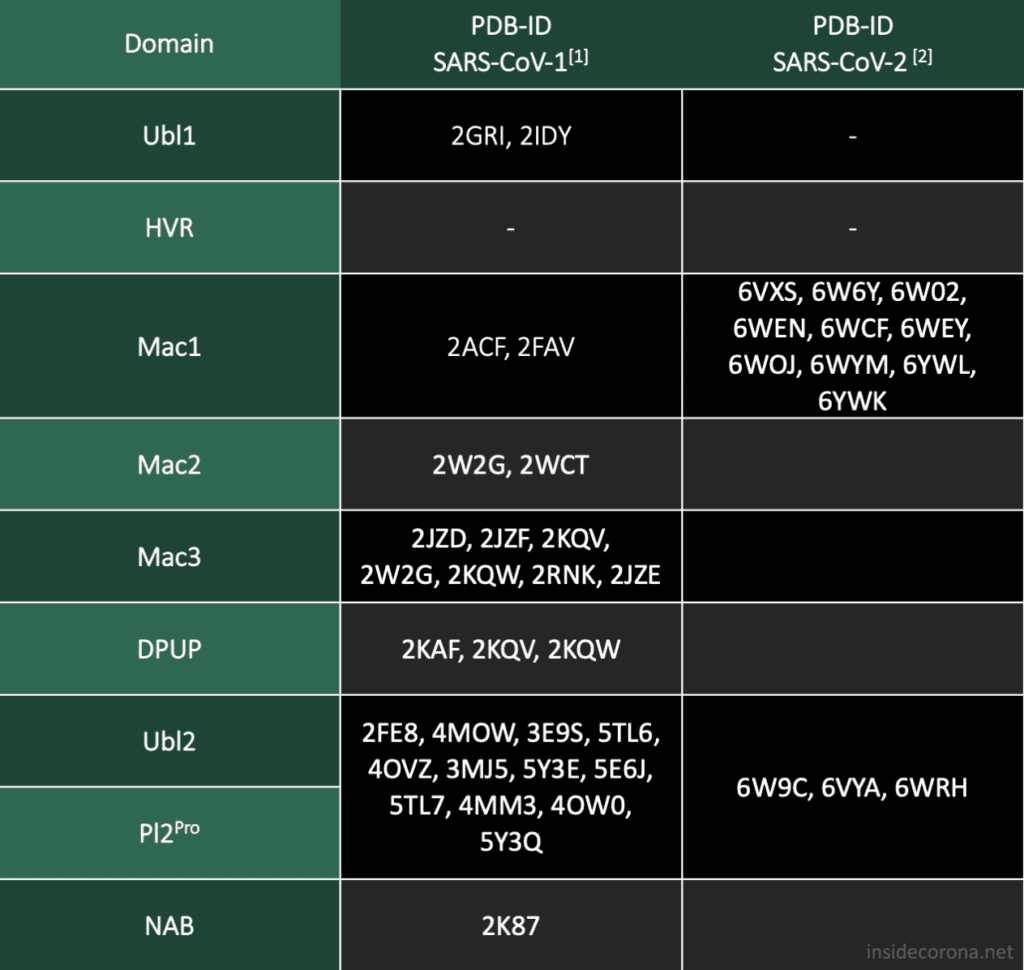
Picture by Kristopher Nolte
Sources
- [1] Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018 Jan;149:58-74. doi: 10.1016/j.antiviral.2017.11.001. Epub 2017 Nov 8. PMID: 29128390; PMCID: PMC7113668.
- [2] Madden T. The BLAST Sequence Analysis Tool. 2002 Oct 9 [Updated 2003 Aug 13]. In: McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2002-. Chapter 16. Available from: http://www.ncbi.nlm.nih.gov/books/NBK21097/
- [3] https://github.com/thorn-lab/coronavirus_structural_task_force
- [4] Benjamin W. Neuman, Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles, Antiviral Research, Volume 135, 2016, Pages 97-107, ISSN 0166-3542, https://doi.org/10.1016/j.antiviral.2016.10.005.
- [5] K.R. Hurst, C.A. Koetzner, P.S. Masters, Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex J. Virol., 87 (2013), pp. 9159-9172
- [6] Kusov Y, Tan J, Alvarez E, Enjuanes L, Hilgenfeld R. A G-quadruplex-binding macrodomain within the "SARS-unique domain" is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015 Oct;484:313-22. doi: 10.1016/j.virol.2015.06.016. Epub 2015 Jul 3. PMID: 26149721; PMCID: PMC4567502.
