Leider steht für diesen Artikel keine deutsche Übersetzung zur Verfügung.
Introduction
It is known as VUI‑202012/01 or B.1.1.7 – the new mutation of the coronavirus Sars-CoV-2. It may be responsible for a sharply increased number of infections in the southeast of England (1), however, the scientific results leading to very strict lockdown measurements in the south of the UK, and travel restrictions across Europe are few and far between. Here, we have compiled what is known up until now.
On mutations
Mutations are normal in the evolution of life – and of viruses. If two similar viruses have infected the same cell, their genomes can become mixed-up, one of the reasons why animal influenza strains are considered so dangerous. This is also called recombination. Mutations can be caused by chemicals, radiation (including UV light) and errors during genome copying. A typical SARS-CoV-2 virus accumulates two amino acid changes per month in its genome — a rate of change about half that of influenza (2). This is because SARS-CoV-2 can repair RNA to some extent. But even so, this natural process led to thousands of mutations since the beginning of the pandemic. If they affected the virus life cycle negatively, that strain may have likely died out - if they did not make a difference or enhanced its chances of survival, it may have persisted.
Many mutations that are observed occur in the spike protein, which both serves to recognize potential host cells but is also what is being recognized by antibodies (i.e., the immune system).
Changes here can be crucial for the survival of the virus (“evolutionary pressure”) as they could significantly alter its affinity to the human receptor ACE2, which the virus uses as gateway to our cells.
What vaccines do
Most, if not all, potential COVID-19 vaccines expose our body to some part of the spike protein, which can be made by the body itself (mRNA vaccines) or carried by a harmless virus instead of SARS-CoV-2 (vector). Our body then produces antibodies which specifically recognize the spike and persist for several months. If we are exposed afterwards to the real virus, the body can recognize it immediately – and the risk of infection is much lower as the immune system swings into action immediately. Earlier this year, the spike mutation D614G (amino acid residue number 614 changing from aspartic acid (D) to glycine (G)) caused quite a stir in the media, and became the predominant form of SARS-CoV-2 (2, 3). However, if and in how far this was caused by natural selection is still debated (3). Another example which triggered an increased media coverage was the mutation Spike Y453F, which originated from infected minks in Denmark (4) and led to a culling of millions of animals. In any case, if we would be vaccinated with a spike protein form that would be different from the one in a virus we encounter later, there is a small chance that the vaccine may be rendered ineffective. This chance is, however, small for SARS-CoV-2, in any case much smaller than for HIV, which famously evaded any attempt to develop a vaccine.

What do we know?
There was a steep rise in infections in the UK recently, as in most other European countries.
A new mutation of the virus has emerged and seems to replace the old version of SARS-CoV-2 (5). Thousands of patients have been found to carry this variant.
This new variant has more mutations at once than expected. These mutations have not observed in this combination before.
The variant has been reported in the UK, the Netherlands, Denmark, Australia and Belgium so far.
What is striking to me as scientist about these findings is one thing in particular: How could the British government find that thousands of people were having the new SARS-CoV-2 variant, instead of the old, if the illness does not look any different? Sequencing samples from each and every patient would be technically very challenging, if not impossible. How could they know? The answer is:
Serendipity
The main PCR test employed in the United Kingdom is Thermo Fisher's TaqPathCOVID-19. This test identifies RNA on three different genome locations: In ORF1ab, nucleotide and spike. Now, it stopped working for the spike portion of the test, while the other two RNAs were still found to be present, which likely prompted scientists to sequence some of the samples in question. And indeed, the new mutant has a deletion of histidine-69 and valine-70, called 69-70del. This permitted easy differentiation of patients with the old SARS-CoV-2 (3 hits) and the new (2 hits) and is the reason why we know so much about the epidemiology of this variant!* It has also to be said that this test is not used as often in other countries, such as Germany, and this could well be the reason why we do not know if and how widespread it is here. In addition, other countries sequence much smaller proportions of virus isolates than the UK, so ongoing circulation of this variant outside of the UK cannot be excluded.
The details of the mutation
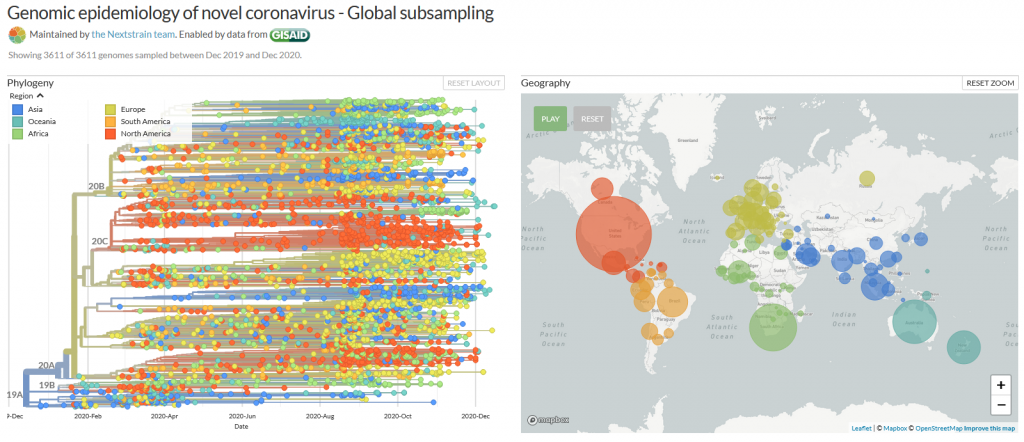
The new variant of SARS-CoV-2 VUI-202012/01 has 14 amino acid changes and three deletions affecting the genes for ORF1ab, spike and ORF8. One of these mutations (N501Y) occurs in the receptor binding domain and could lead to an increased binding affinity to the human ACE2. The 69-70 deletion has likely an immunological role and is the reason this mutant was detected so widely, as this RNA location is used for PCR tests. Another interesting mutation is the P681H, which is next to a furin cleavage site that has a biological significance in membrane fusion. These mutations could be responsible for the increased transmissibility. The effects of the other mutations aren’t fully investigated yet. Here is a list of the mutations which have been observed in the VUI‑202012/01 or B.1.1.7 variant:
| T1001I in gene ORF1ab | |
| A1708D in gene ORF1ab | |
| I2230T in gene ORF1ab | |
| SGF 3675-3677 deletion in gene ORF1ab | |
| A1708D in gene ORF1ab | |
| HV 69-70 deletion in spike | The 69-70 deletion on the spike protein is a re-occurring mutation that has shown to often co-occur with other amino acid changes in the RBD (6, 7). (1) Evasion to the human immune response and in association with other receptor binding domain changes (1) (2) Immunological role (8) (3) Leads to diagnostic failures which permit detection (see above, "Serendipity") (4) Associated with immune escape in immunocompromised patients (9(8)) Furthermore, the 69-70 deletion arose in multiple unrelated lineages and is associated with the evasion of the immune response (9). It is being hypothesized that this mutation undergoes a strong positive selection when exposed to convalescent plasma therapy in an immunocompromised human host (7). |
| Y144 deletion in spike | Deletion in the spike N-terminal domain (9) |
| N501Y in spike | One of six key contact residues in the spike receptor binding domains, this mutation leads to an increasing binding affinity to human and murine ACE2 (1). |
| A570D in spike | Mutation located at the spike receptor binding domain (10) |
| P681H in spike | The P681H mutation is located directly next to the furin cleavage site. It is one of the four residues which are insertions when compared to closely related coronaviruses, creating a furin cleavage site in the spike protein between the spike S1 and S2 domains. This prompts the entry of the virus into respiratory epithelial cells as well as the transmission in animal models (1) The S1/S2 furin cleavage site of SARS-CoV-2 is not found in closely related coronaviruses and has been shown to promote entry into respiratory epithelial cells and transmission in animal models (9) |
| T716I in spike | Mutation in in the S2 domain |
| S982A in spike | Mutation in in the S2 domain (10) |
| D1118H in spike | Mutation in in the S2 domain (8) |
| Q27 stop in ORF8 | The Q27stop mutation in the ORF8 leads to the truncation of the ORF8, and as it only consists of 121 amino acids, the consequence might be a loss of function. These and the other mutations could be responsible for the increased transmissibility of the B.1.1.7 variant. In any case, this mutation truncates the ORF8 protein at residue 27 or renders it inactive which allows further downstream mutations to accrue. (1) |
| R52I in ORF8 | |
| Y73C in ORF8 | |
| D3L in nucleocapsid | |
| S235F in nucleocapsid |
Why were there so many mutations at once?
This could be a result of prolonged or chronical SARS-CoV-2 infections as study of these infections reveal unusually large numbers of nucleotide changes and deletion mutations and often high ratios of non-synonymous changes. In addition to this, convalescent plasma treatment can cause intra-patient virus genetic diversity (11).
What does the new mutation mean in terms of impact and epidemiology?
There was an increase in cases with the new strain in total and in
proportion to the old (1). What does that mean for us?
This is what the internet says:
The COVID-19 genomics UK consortium (COG) reports about a “priority set of SARS-CoV-2 Spike mutations that are of particular interest based on potential epidemiological significance in the UK and/or biological evidence based on the literature or unpublished work.” (9)
The New and Emerging Respiratory Virus Threats Advisory Group of the British government (NERVTAG) discussed the new variant on Friday and concluded that its growth rate is higher by 67-75% and that this is likely due to a selective advantage. “In summary, NERVTAG has moderate confidence that VUI-202012/01 demonstrates a substantial increase in transmissibility compared to other variants.” (12) This is very likely the source of Boris Johnson’s claim to this strain being “70% more infectious”.
The English government writes that PHE (Public Health England) „is working with partners to investigate and plans to share its findings over the next 2 weeks. There is currently no evidence to suggest that the variant has any impact on disease severity, antibody response or vaccine efficacy. High numbers of cases of the variant virus have been observed in some areas where there is also a high incidence of COVID-19. It is not yet known whether the variant is responsible for these increased numbers of cases.” (13)
Conclusion
From this, we conclude that the British government, and we, do not know yet. It has not been conclusively shown that the new variant is more infectious (likely), has an easier time to evade the host immune system or if the vaccine will be less effective against it (very unlikely). The epidemologic model which predicts a higher tranmissability has still to be published, the science is still in the making. Tests of vaccines against the new variant are ongoing and will take a few weeks. There is yet little evidence that this new variant poses a significantly bigger threat than others - or to the contrary.
Acknowledgements
While I am listed as author of this article, it could not have been written without the help and research by Pairoh Seeliger, Lea von Soosten, Luise Kandler, Erik Nebelung and Oliver Kippes who all helped in this.
I would also thank Nicolai Wilk from Thermo Fisher Scientific who quickly responded to my questions about their test.
The title picture shows mutation cards from the game Pandemic Expansion: On the Brink by Z-Man Games.
- *The 69-70del mutation is predominantly observed in B.1.1 (including B.1.1.7), B.1.258, and the cluster 5 variant lineages of SARS-CoV-2.
References
- 1.A. Rambaut, Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. virological.org (2020), (available at https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563).
- 2.E. Callaway, The coronavirus is mutating — does it matter? Nature, 174–177 (2020).
- 3.L. Zhang, C. B. Jackson, H. Mou, A. Ojha, H. Peng, B. D. Quinlan, E. S. Rangarajan, A. Pan, A. Vanderheiden, M. S. Suthar, W. Li, T. Izard, C. Rader, M. Farzan, H. Choe, SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun (2020), doi:10.1038/s41467-020-19808-4.
- 4.ECDC, Detection of new SARS-CoV-2 variants related to mink. www.ecdc.europa.eu (2020), (available at https://www.ecdc.europa.eu/sites/default/files/documents/RRA-SARS-CoV-2-in-mink-12-nov-2020.pdf).
- 5.ONS UK , Percentage of COVID-19 cases that are positive for ORF1ab and N genes. www.ons.gov.uk (2020), (available at https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/adhocs/12690percentageofcovid19casesthatarepositivefororf1abandngenes).
- 6.R. M. Dawood, M. A. El-Meguid, G. M. Salum, K. El-Wakeel, M. Shemis, M. K. El Awady, Bioinformatics prediction of B and T cell epitopes within the spike and nucleocapsid proteins of SARS-CoV2. Journal of Infection and Public Health (2020), doi:10.1016/j.jiph.2020.12.006.
- 7.S. A. Kemp, D. A. Collier, R. Datir, S. Gayed, A. Jahun, M. Hosmillo, I. A. Ferreira, C. Rees-Spear, P. Mlcochova, I. U. Lumb, D. Roberts, A. Chandra, N. Temperton, K. Sharrocks, E. Blane, J. A. Briggs, K. G. Smith, J. R. Bradley, C. Smith, R. Goldstein, I. G. Goodfellow, A. Smielewska, J. P. Skittrall, T. Gouliouris, E. Gkrania-Klotsas, C. J. Illingworth, L. E. McCoy, R. K. Gupta, Neutralising antibodies drive Spike mediated SARS-CoV-2 evasion (2020), , doi:10.1101/2020.12.05.20241927.
- 8.K. Kupferschmidt, Mutant coronavirus in the United Kingdom sets off alarms, but its importance remains unclear. Science (2020), doi:10.1126/science.abg2626.
- 9.COG, COG-UK update on SARS-CoV-2 Spike mutations of special interest Report 1. https://www.cogconsortium.uk (2020), (available at https://www.cogconsortium.uk/wp-content/uploads/2020/12/Report-1_COG-UK_19-December-2020_SARS-CoV-2-Mutations.pdf).
- 10.S. Kemp, W. Harvey, R. Datir, D. Collier, I. Ferreira, A. Carabelii, D. L. Robertson, R. K. Gupta, Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion ΔH69/V70 (2020), , doi:10.1101/2020.12.14.422555.
- 11.ECDC, Threat Assessment Brief: Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. www.ecdc.europa.eu (2020), (available at https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-rapid-increase-sars-cov-2-variant-united-kingdom).
- 12.NERVTAG, NERVTAG meeting on SARS-CoV-2 variant under investigation VUI-202012/01. https://khub.net (2020), (available at https://khub.net/documents/135939561/338928724/SARS-CoV-2+variant+under+investigation%2C+meeting+minutes.pdf/962e866b-161f-2fd5-1030-32b6ab467896?t=1608470511452).
- 13.PHE, PHE investigating a novel variant of COVID-19 . www.gov.uk (2020), (available at https://www.gov.uk/government/news/phe-investigating-a-novel-variant-of-covid-19).
Für diesen Beitrag exisitiert leider keine deutsche Übersetzung.
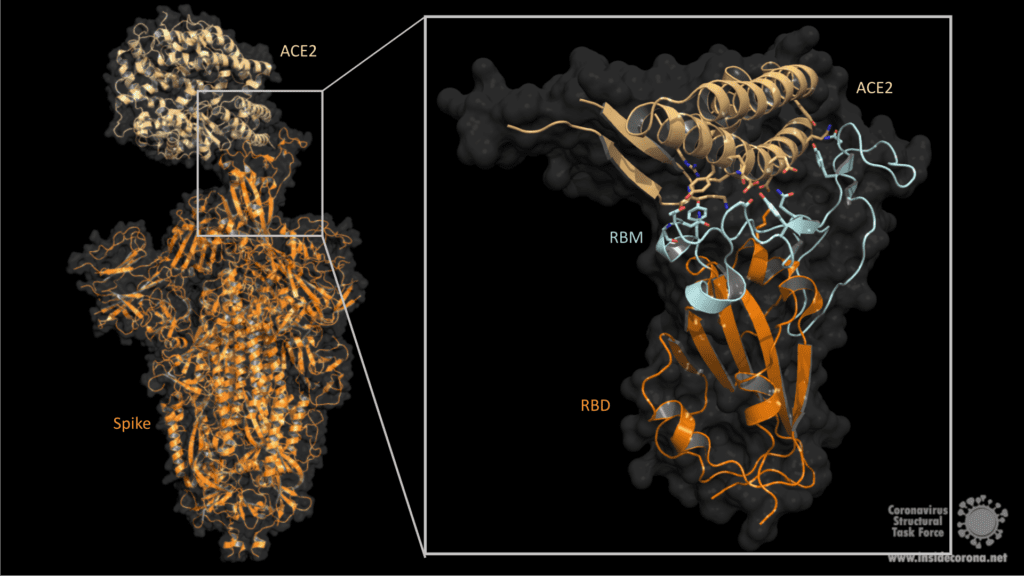
COVID-19 is caused by the new coronavirus SARS-CoV-2. This virus has a characteristic virus hull featuring surface proteins which are commonly called “spikes”. Protruding from the viral hull like “spikes of a crown”, they give the coronavirus its name (corona = crown). These proteins make the first contact with human cells and are akin to keys that use a human receptor called “angiotensin-converting enzyme2” (ACE2) as a backdoor to gain access to and infect the cell.
1. Fuction of ACE2
ACE2 is a membrane protein which is anchored in the human cell membrane of epithelial cells. This type of cells can be found on the surface of lung, intestine, heart and kidney tissue. As a type I membrane protein, its primary function is to take part in maturation of angiotensin, a peptide hormone which controls vasoconstriction and blood pressure. ACE2 can be compared to a lock which can be unlocked by the coronavirus spike protein. The virus can then enter the cell and hijack its functions to reproduce itself, thus causing the Covid-19 infection which poses a serious danger to humanity, especially for older people and people with pre-existing conditions. For this reason, one approach to combating SARS-CoV-2 is to target and inhibit the spike to prevent infection. In order to do so, knowledge of the structural features of the spike and its interaction processes with ACE2 are indispensable. (Further information about how macromolecular structures are visualized can be found on our homepage: https://insidecorona.blogs.uni-hamburg.de/visualizing-macromolecular-structures/)
2. Spike: Structure and Fusion Mechanism
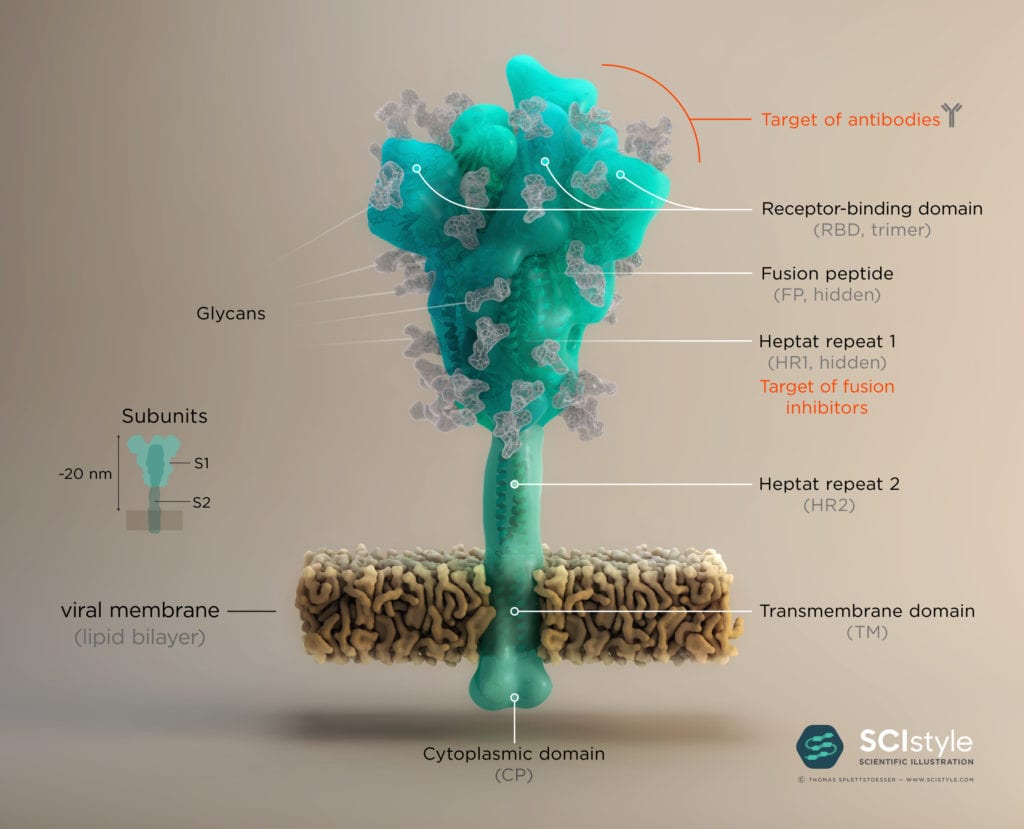
The Spike protein has a trimeric shape comprising three identical monomeric structural elements. Each of these monomers can fold out akin to a modern car key with a fold-out key element with specific teeth on its surface. This fold-out key element is the so-called “receptor binding domain” (RBD). The spike can only interact with ACE2 when its RBD is in a folded-out position, exposing its teeth, or “receptor binding motive” (RBM). As the name suggests, it comprises a motive of different amino acids which then can bind and unlock the ACE2 receptor. This key lock mechanism triggers a cascade of events initiating fusion with the host cell. First, protein scissors are recruited to the binding site. These scissors (furin & transmembrane serine protease 2) cleave the spike protein for subsequent activation. The active spike molecule then rearranges itself to form a long structural “hook” (formed of HR1/ HR2 and FP see Fig.2) that brings the epithelial cell and viral cell membrane into close proximity for fusion. Once the fusion is completed, the path for the virus is clear to transfer its genome encoded in ribonucleic acid (RNA) into the host cell. This successful transfer then enables the virus to multiply itself and finally spread from cell to cell, causeing Covid-19 in its wake.
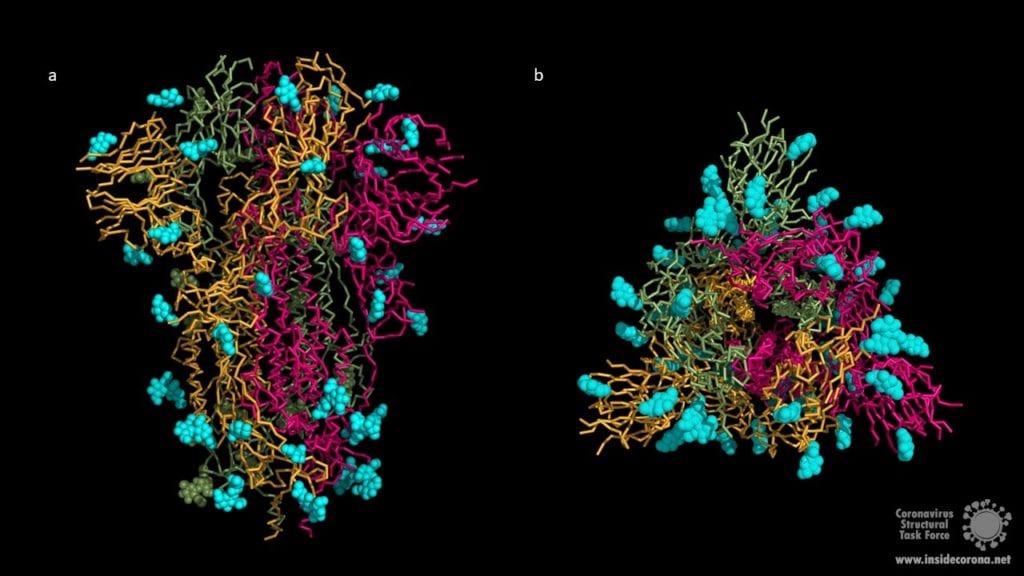
3. Evading the Immune System with Carbohydrate Chains
The human immune system normally recognizes the surface proteins of foreign organisms such as viruses or bacteria and reacts with an immune response to combat them. Spike proteins are such surface proteins but because of structural peculiarities, the coronavirus evades both the innate and the adaptive human immune system. The secret of these structural peculiarities are the N-glycans. These are long carbohydrate chains which sit on spike’s surface. Each spike comprises 66 N-glycans forming a protective shield around the protein. Hence the human immune system has problems recognizing spikes and identifying the coronavirus as an enemy.
The COVID 19 pandemic has a massive impact on our lives, our health and the global economy. Scientists around the world are trying to develop new drugs to combat the virus. Since the spike plays a critical role in the infection process, it is a prime target for drug development against the pandemic. One drug approach to inhibit the interaction between spike and the ACE2 receptor is to cap the spike protein using antibodies. Antibodies are proteins, normally produced by the human immune system to fight viruses. The idea is to treat patients with antibodies that cap the RBD of spike, thus preventing interactions with ACE2. This would lead to a nonfunctional spike, blocking the coronavirus from entering the cell (The key would no longer fit the lock). Another approach includes the development of small molecules that target and inactivate the protein scissor transmembrane serine protease 2 (see chapter 2), as the spike’s functionality depends on its cleavage activity. Since the spike protein decorates the virus hull, it could even be part of a potential vaccine. For this reason, the spike protein could also become the key in the molecular fight against COVID-19.
