Accessory proteins are not essential for viral replication in vitro and have been studied less than the other proteins. They have been named based on their location within the SARS-CoV genome. Sequences of RNA are found to encode different proteins through varied Open Reading Frames (ORFs). This means that the starting point on the RNA for proteins can vary, resulting in various proteins being produced by the same sequence of RNA. For example, RNA can code simultaneously for ORF9b and Nucleocapsid, depending where you start.
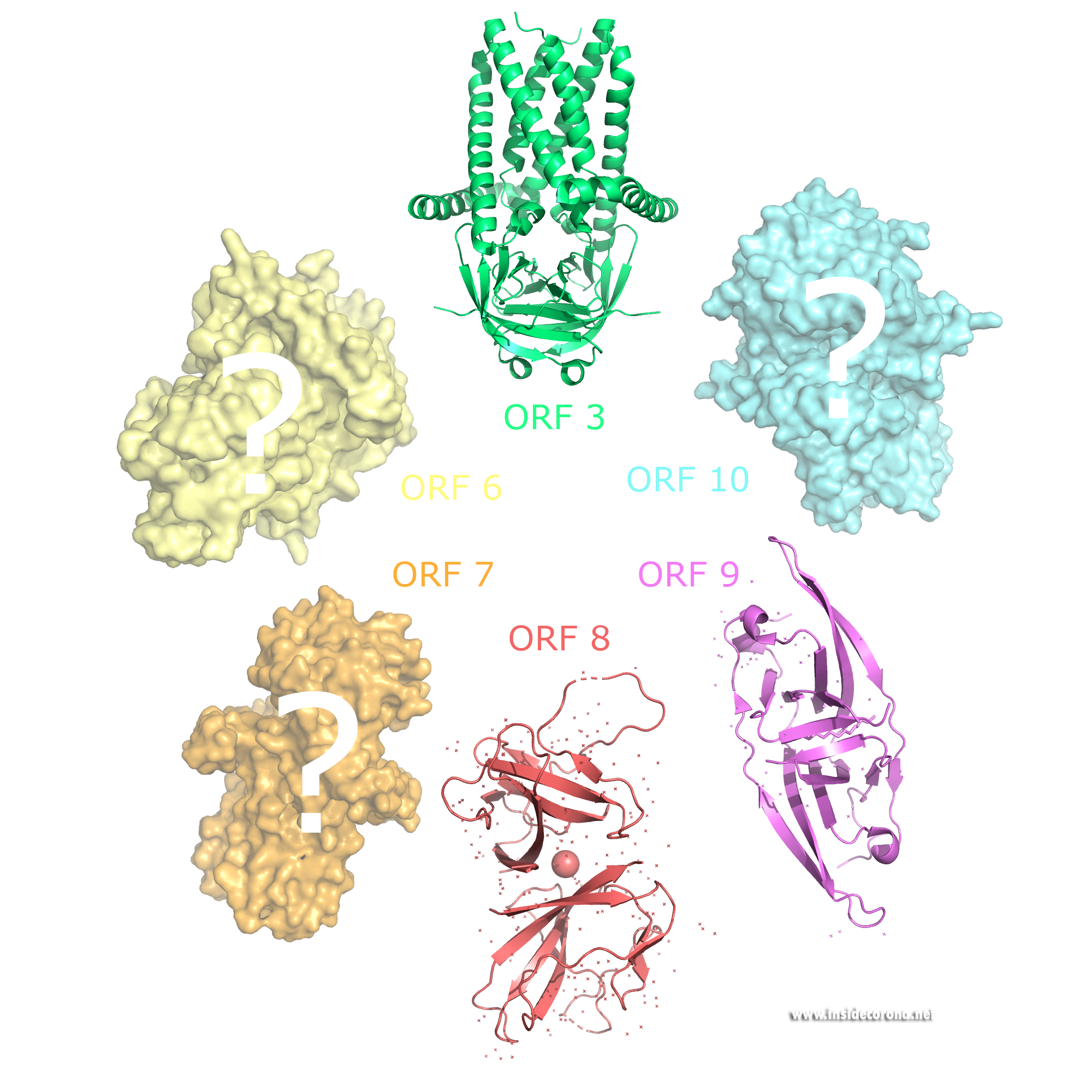
ORF3 is only 22 amino acids long in SARS-CoV-2, but 154 amino acid residues in SARS-CoV.
It has been shown to suppress interferon, an important part of the immune response in the host cell. In some patients in Peru, the SARS-CoV-2 protein appears longer, and scientists demonstrated that the longer ORF3 is the better it can suppress the immune system. This makes the Peru subtype rather worrying.
ORF3 also seems to play a crucial role in making the host cell permeable to the excretion of virions and may interact with the spike, membrane, and envelope proteins.
As a fun aside, the structure of ORF3a looks like a pig wearing a hat. (Compare: https://www.rcsb.org/structure/6XDC)
ORF6 also stops interferon signals, which helps the virus to infect healthy cells.Unlike ORF3 it seems to be a non-essential player in virus replication. Studies have shown that ORF6 could also be an inhibitor for the expression of essential genes to establish resistance mechanisms in the cell. No atomic structure of ORF6 has been determined so far.
ORF7 has been suggested to induce cell death, which may contribute to the severity of COVID-19 infections in humans. Research also suggests it limits the host cell’s supply of tetherin, which a cell’s use to retain excreted virions on its surface to protect surrounding cells for being infected too.
ORF8 carries sugar chains (glycosylation) on its surface which protects it from degradation. It is found in both the cytoplasm and in the endoplasmic reticulum. It has the lowest sequence identity between SARS-CoV and SARS-CoV-2 of any protein found in both viruses. There has been evidence of ORF8 directly interacting with major histocompatibility complex molecules of infected cells, which are an important part of the immune system, leading to MHC-I lysosomal degradation. This results in the limitation of cytotoxic T cell destroying infected cells. An atomic structure of ORF8 has previously been determined.
The gene for accessory protein ORF9b is found within an alternative open reading frame in the nucleocapsid gene. Structures of this protein are currently available for both SARS-CoV and SARS-CoV2. The novel fold of this protein forms a dimeric β-fold with hydrophobic interface with a cavity that is thought to bind lipids. It has been suggested to be membrane attached and could have a minor structural role in the assembly of the virion.