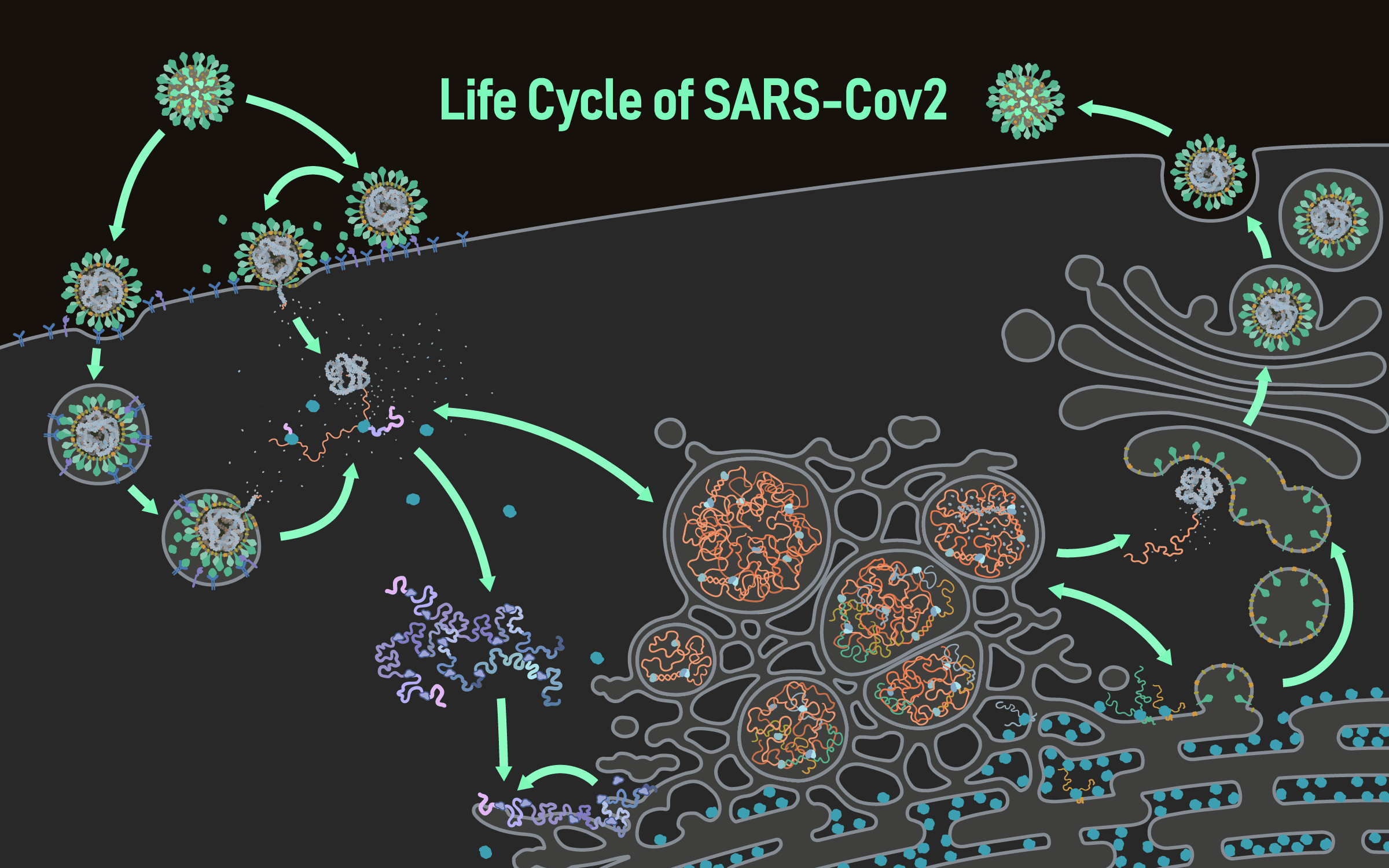
The life cycle of coronavirus
Please click on one of the orange numbers to learn more about this part of the reproductive cycle!
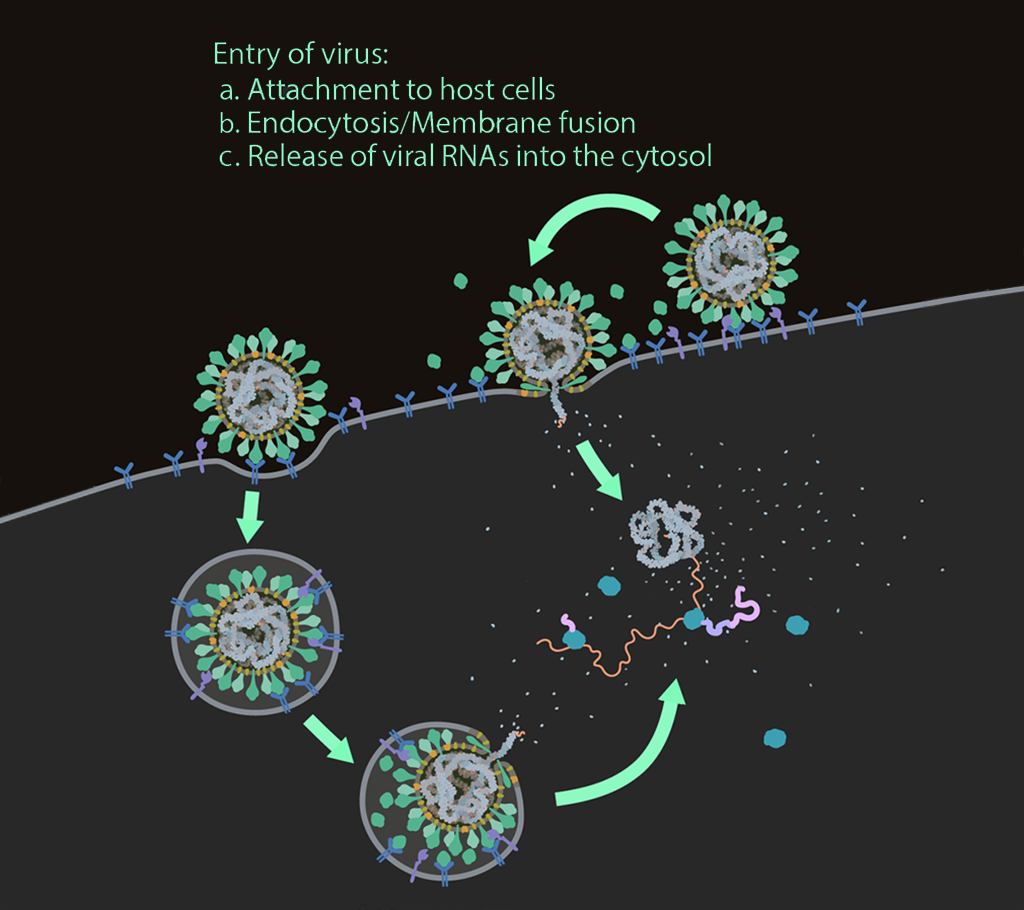
Entry of virus
Infection with SARS-CoV-2 starts upon entry into the host cell via glycosylated spike proteins, which attach to the host cell receptor angiotensin-converting enzyme 2 (ACE2).
After attachment, SARS-CoV-2 enters the host cell via endocytosis or fusion at the plasma membrane, which is driven by protease cleavage and activation.
SARS-CoV-2 then releases its genomic RNA (positive sense) into the cytosol of the host cell for viral RNA and protein synthesis.
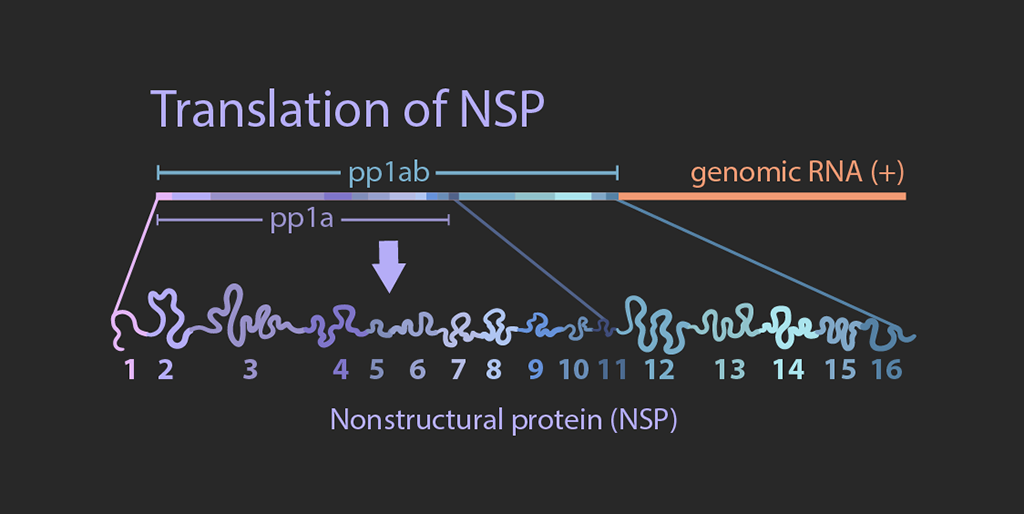
Translation of NSPs
The viral genome is about 30kb long. The 5′ two-thirds of the genome encodes polyproteins pp1a and pp1ab, while the remaining one third consists of genes encoding structural proteins including envelope (E), membrane (M), nucleocapsid (N), and spike (S) proteins.
In the cytosol, viral genomic RNAs are translated by the host ribosome machinery into polyproteins pp1a and pp1ab, which are then proteolytically processed.
More about RNA and nucleocapsid
More about NSP1 which selectively can block ribosomes
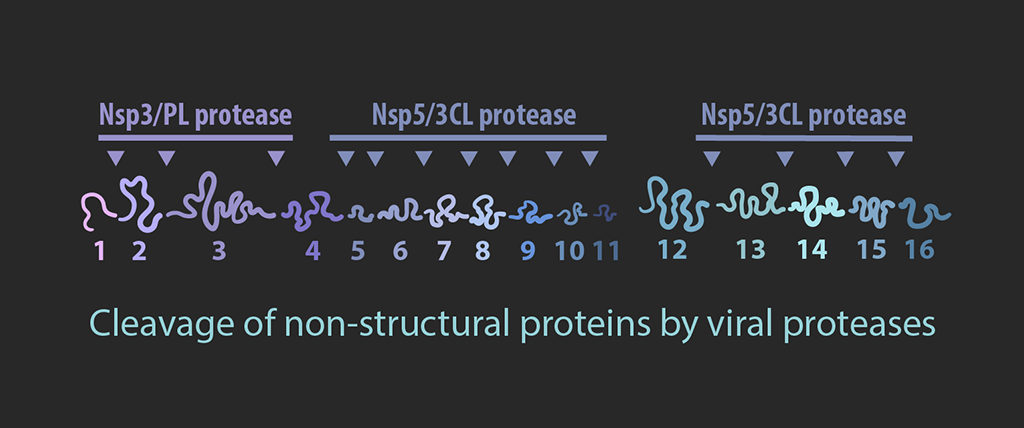
Cleavage
Polyproteins pp1a and pp1ab are cleaved into smaller non-structural proteins (nsps) by viral proteases nsp3 and nsp5.
Nsp3 is a membrane protein that is also involved in the formation of double membrane vesicles (DMVs) with nsp4 and nsp6. Nsp12 is a RNA dependent RNA polymerase, which forms the core structure of the replicase-transcriptase complex with nsp7 and nsp8. Nsp14 is an exoribonuclease that has both proofreading and capping activities, which is facilitate by nsp10. Nsp16 and the helicase, nsp13, are also involved in formation of the RNA cap.
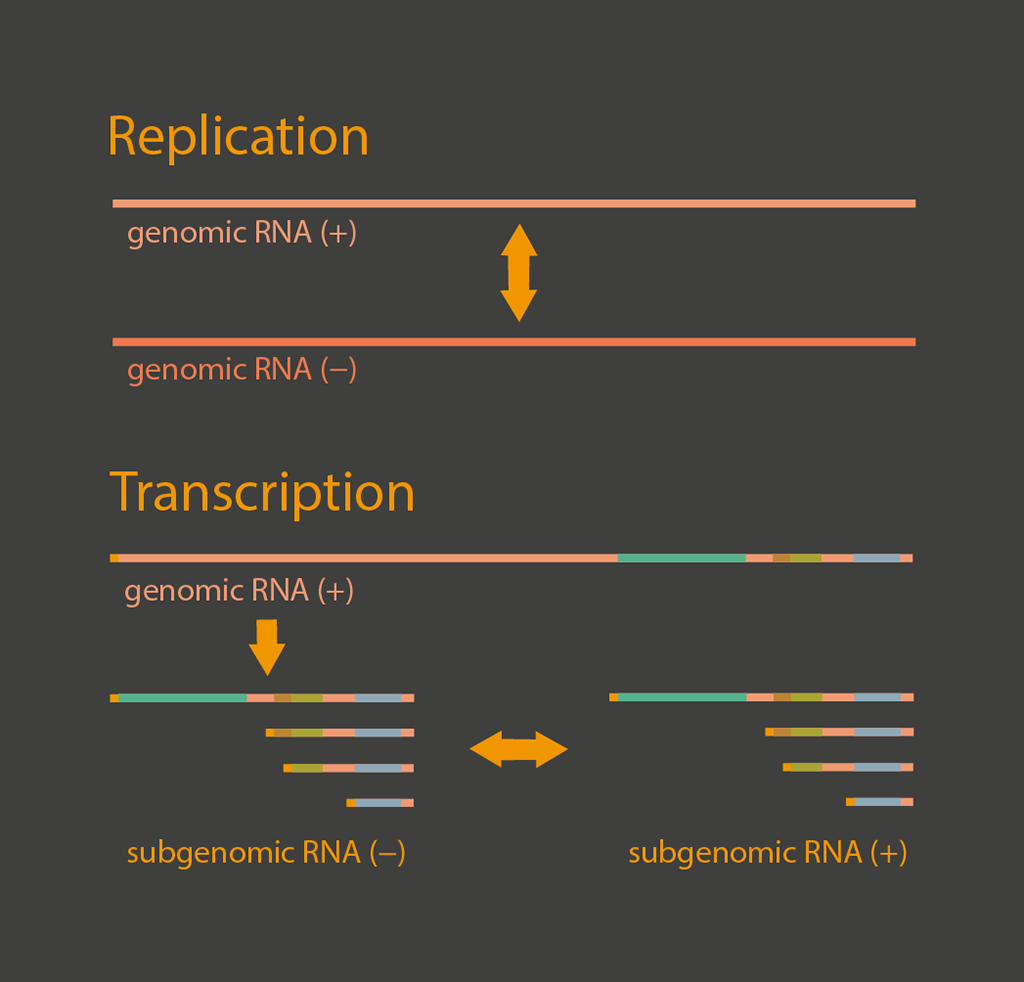
Replication and transcription
Viral RNA synthesis occurs in double-membrane vesicles (DMVs) that are modified from intracellular membranes.
Within the DMV, viral RNAs are replicated to produce full-length negative stranded RNAs that can be used for synthesis of genomic positive sense RNAs and subgenomic negative stranded RNAs through discontinuous transcription. The subgenomic negative stranded RNAs are further transcribed into subgenomic positive stranded RNAs. These positive stranded RNAs are exported into the cytosol through molecular pores that span the double membrane of DMVs for translation of viral proteins.
More about the RNA-Polymerase"
NSP9, EndoRNase and the methyltransferases may be somehow involved - but their role is unclear.
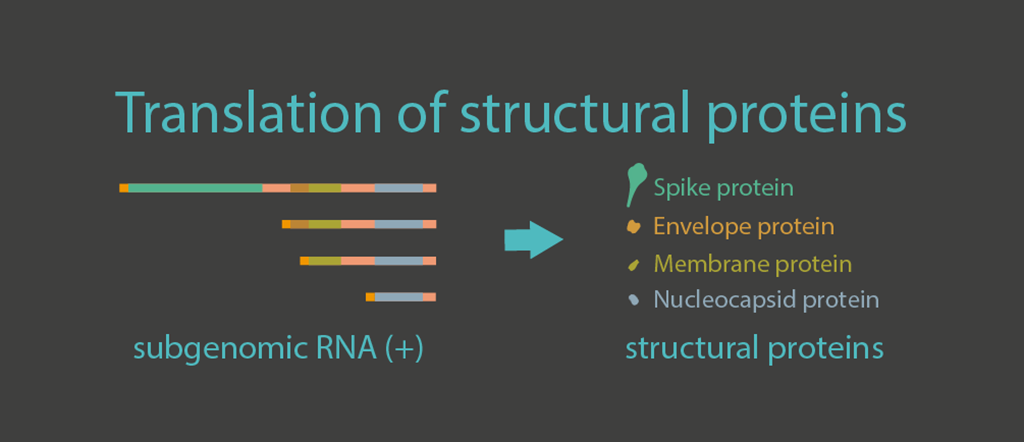
Translation of structural proteins
The 3′ one-third of the viral genome consists of genes encoding structural proteins including envelope (E), membrane (M), nucleocapsid (N), and spike (S) proteins. The transcripts of these structural proteins are generated within double-membrane vesicles and later exported to the cytosol and ER, where they are translated into proteins. E and M proteins are small membrane proteins that are essential for virus assembly and budding. N proteins condense the viral genomic RNA so it can be packaged into the virion. The S protein plays a key role in the receptor recognition and cell membrane fusion process.
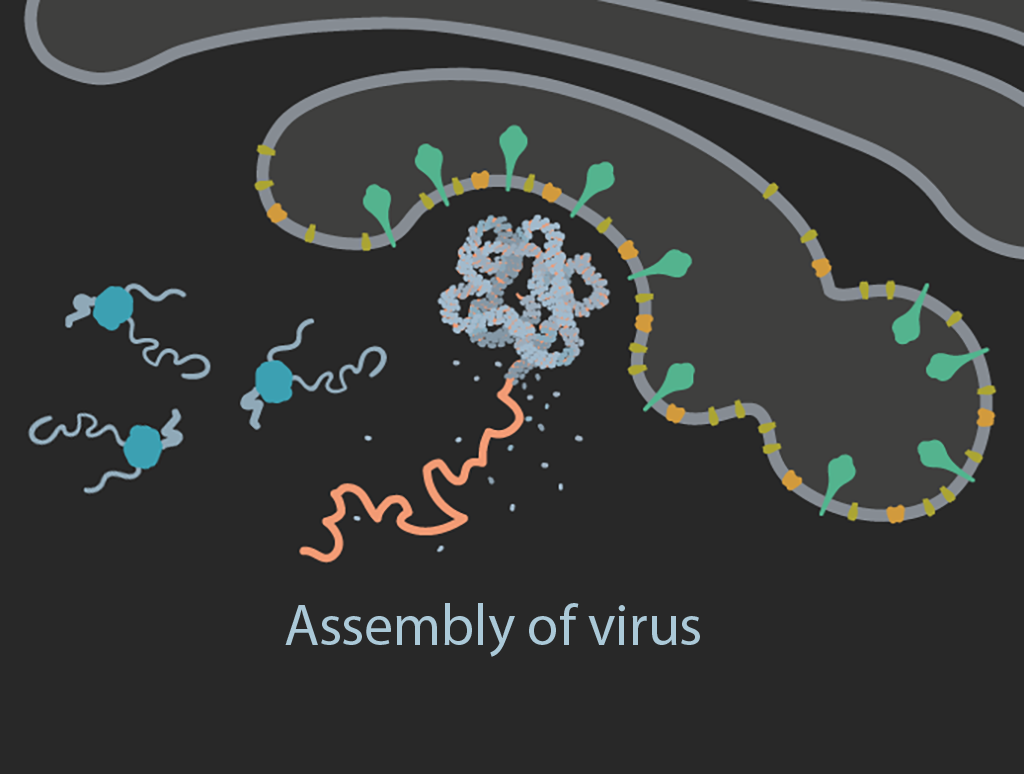
Assembly
Viral genomic positive stranded RNAs and N proteins assemble into nucleocapsids that bud into the endoplasmic reticulum Golgi intermediate compartment (ERGIC). The structural proteins located on the ERGIC membranes are incorporated into the envelope as the virion forms.
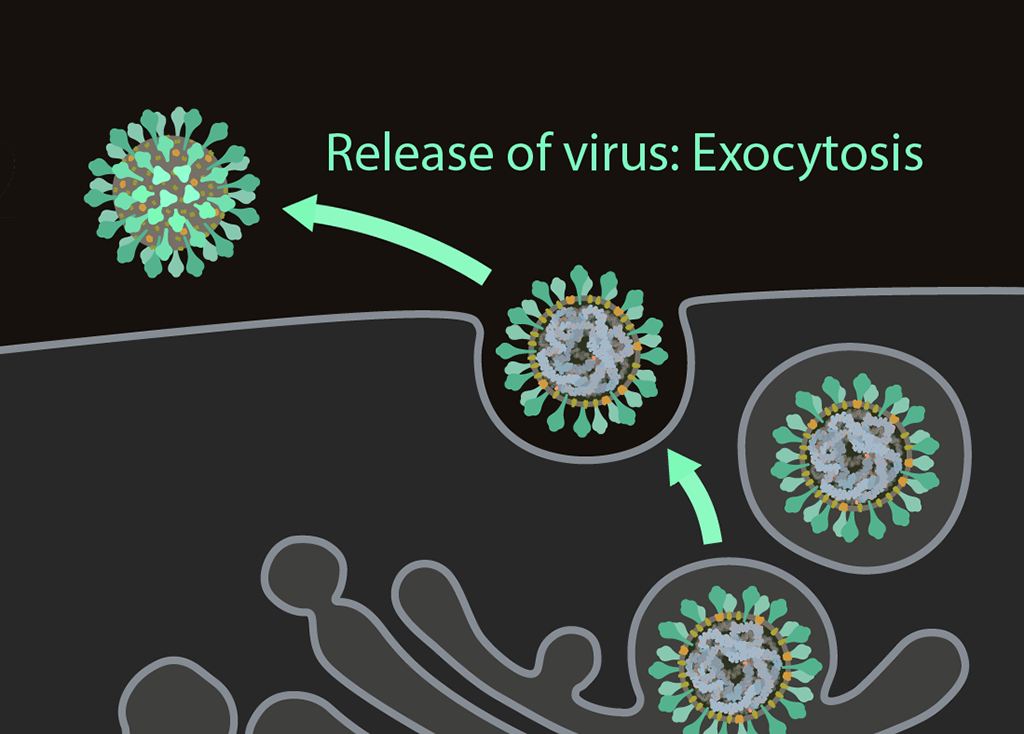
Exocytosis
Once have completed their life cycle, the viruses are ready to be released from the cell through exocytosis for further transmission.
