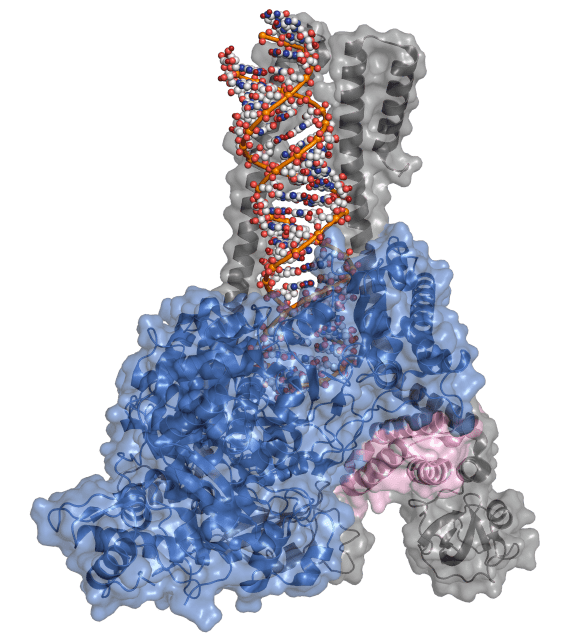
(Also known as: RNA-dependent RNA polymerase, RdRp complex, nsp12, nsp7, nsp8)
SARS-CoV-2 uses single-stranded RNA to encode its proteins, and hence belongs to a class of "single-stranded RNA viruses".
The virus requires a special protein to copy this rather large RNA genome, called an “RNA-dependent RNA polymerase”. This protein uses the viral RNA as a template to make a new copy of RNA, by stringing single ribonucleotides together like beads on a string.
RdRp (also called nsp12) enlists the help of several other proteins – nsp7, nsp8, nsp10, and nsp14.
Nsp7 and nsp8 work together to provide a ramp (called “sliding-poles”) for newly-made RNA to leave the polymerase smoothly. This may seem unnecessary, but the polymerase can’t make new RNA without it!
Nsp10 and nsp14 work together to proofread the newly-made RNA, cutting out any mistakes, which allows SARS-CoV-2 to have such a huge genome.
Together, these proteins form a complex machine that enables the virus to make many high-fidelity copies of itself.