1. Introduction:
With SARS-CoV-2 infections and related death rates continuing to rise worldwide and new variants emerging, the virus is still a great and present danger. Although we have gathered significant knowledge and the first vaccinations have started, new mutations can still set our efforts back and possibly make the virus even more potent. Thus, searches for new treatments are of paramount importance. The nucleocapsid structural protein, or N-Protein, could serve as another drug target.
The nucleocapsid’s main function is to protect the genomic RNA by packaging it into a ribonucleoprotein complex (RNP). Apart from this, the protein has other functions essential for the viral life cycle. It is involved in virion assembly, viral RNA synthesis, transcriptional regulation of genomic RNA, and translation of viral proteins1.
2. Structure
The SARS-CoV-2 nucleocapsid is an RNA-binding protein separated into five domains. Three of the domains are intrinsically disordered, meaning they are challenging for conventional structural characterization . Two of these intrinsically disordered regions (IDRs) are located at the N- and C-terminus of the protein, and the third acting as a linker between the two structured domains. Not much is known about the IDRs: their transient structural details are mostly predictions from molecular simulations2. The other two domains, the RNA-binding domain and the Dimerization domain (DMD), are well organised and their structures have been determined by X-Ray diffraction and NMR.

Disordered Domains:
The Linker
Flexible linkers contain a large number of polar and charged amino acids. The resulting electrostatic repulsion and lack of a stabilizing hydrophobic core prevents a well-structured conformation resulting in the disorder. Experiments have shown that the Linker region of SARS-CoV-2 nucleocapsid incorporates such polar regions which are repelled by the neighbouring folded domains. The Linker contains a positively charged serine and arginine rich motif which likely functions as a phosphorylation site for a direct interaction with RNA, M (membrane) protein, and Nsp31,2. Simulations reveal that the Linker does not often adopt helical conformation as they are transient, but it may contribute to oligomerization or act as a recognition motif for the binding of other proteins. Intrinsically disordered regions, in general, are thought to be involved in a number of regulatory functions including modulation of transcription, translation, post-translational modifications such as phosphorylation, and cell signalling, often through ordering when in contact with another protein domain1.
The disordered N- and C-terminal domains
The N- and the C-terminal regions of the nucleocapsid protein are also disordered but have several regions which also may form transient helices2. The N-terminal conformation is significantly affected by the neighbouring folded RNA binding domain. Electrostatic interactions with the RNA binding domain are proposed to cause a repulsion of the positively charged N-terminal domain from its positive surface of the and an attraction to the slightly negatively charged parts2.
The other disordered tail, the C-terminal domain, interacts with the neighbouring folded dimerization domain, competing with intradomain interactions2.
Folded domains:
The RNA-binding domain and dimerization domain are well-organised folded domains. They make up 257 of the 422 residues in nucleocapsid. All five domains, nevertheless, have been proposed to be involved in RNA-binding1.
The RNA-binding domain
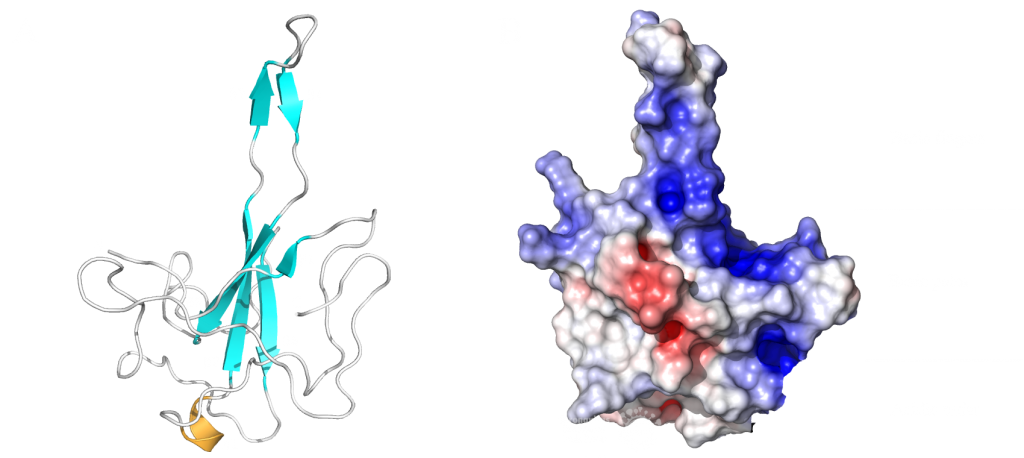
This domain mainly interacts through residues in a positively charged β-hairpin and the so-called palm region. It is rich in aromatic and basic residues that are folded into a right-hand-like shape with a protruded basic finger, a basic palm, and an acidic wrist (see Fig.1 B). Crystal structures from SARS-CoV-2 show two right-handed loops that surround the β-sheet core in a sandwiched structure. The β-sheet core consists of four antiparallel β-strands, a short 310 helix in front of the β2 strand and a protruding β hairpin that is located between the β2 and β5 strands (see Fig. 1 A). The structural basis for RNA binding by nucleocapsid is not yet known but comparisons with the less dangerous virus type HCoV-OC43 suggest a unique potential RNA binding pocket beside the β-sheet core3,4.
The dimerization domain (DMD)
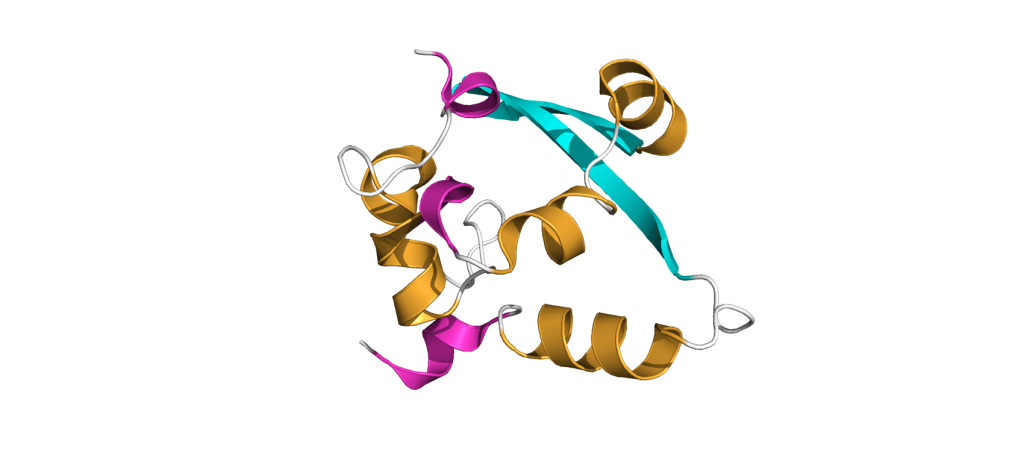
The dimerization domain (DMD) is only stable when several nucleocapsid molecules form a dimer or oligomer. Its structure consists of three 310 -helices, five α-helices and two antiparallel β-strands, which create a β-hairpin. This β-hairpin together with the other parts of the domain form a shape that is like the letter “C”. Two domains form a tight homodimer with a rectangular slab shape, the β-hairpins from each N-Protein are at one side and the helices at the opposite side. The dimer is stabilized through hydrogen bonds and hydrophobic interactions. It is possible that the DMD has RNA binding activity, experiments showed that the amount of free RNA from SARS-CoV 2 is decreased if DMD proteins are added4,5.
The PDB currently has 22 structures that picture the RNA binding domain and the dimerization domain. The structures: 7ACT and 7ACS are particularly interesting because they are the only structures that are in complex with RNA. The RNA binding domain is also a potential inhibitor target and a subject of inhibitor Studies6. The dimerization domain has structures that show the domain as a monomer and a dimer. There is no structure of the whole protein in the PDB yet.
3. Ribonucleoprotein Complex (RNP):
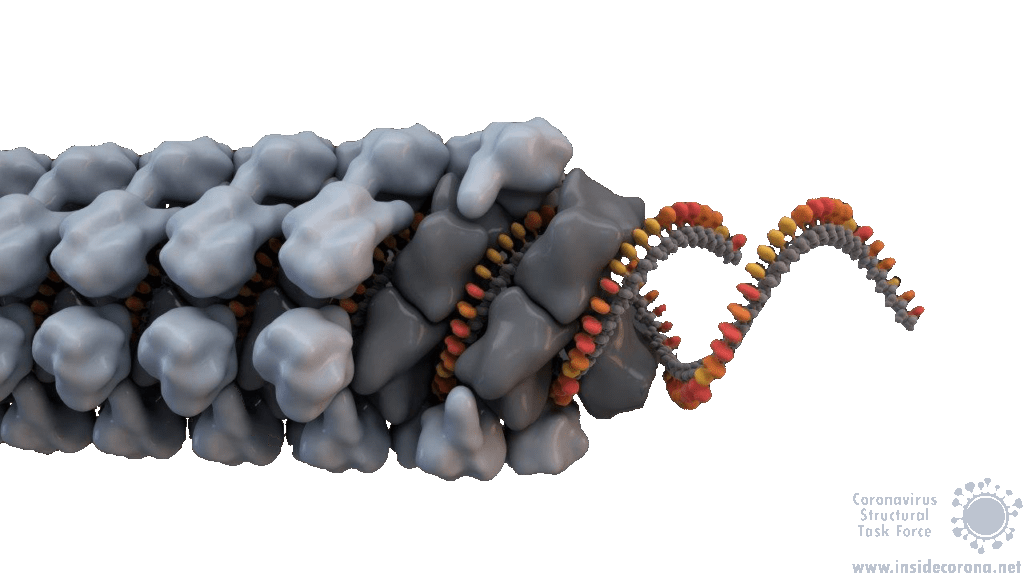
In order to package the viral RNA genome, the nucleocapsid binds the RNA via the RNA binding domain in order to form a long, flexible, helical ribonucleoprotein complex1. Two key functionalities are necessary for this process: The nucleocapsid must interact with the nucleic acid, which is preferentially mediated by GGG motifs from the leader RNA sequences7 and the nucleocapsids need the ability to oligomerize. They interact with the RNA at multiple sites through specific (sequence dependent) and non-specific (sequence independent) binding. Little is known about specific binding to the RNA, but nonspecific binding is likely to involve interactions between the negatively charged phosphate backbone of the RNA and the positively charged groove formed by the residues 248-280 of the N protein. It seems also clear that the nucleocapsid helps RNA folding1. The helical RNPs consist of coils 9 – 16 nm in diameter with a hollow interior 3 – 4 nm wide. It is frequently twisted upon itself and most of the RNPs are supercoiled into compact intertwined structures1. New cryoelectron tomography analysis of SARS-CoV-2 revealed another potential structure of the RNP, this structure is described like ‘beads on a string’ that links RNPs together, and more research is urgently needed[16]. In addition to this, the exact mechanisms of RNA protection through the nucleocapsid are still unknown1.
4. Functions:
Nucleocapsids are multifunctional proteins necessary for the viral life cycle. The main function of the nucleocapsids is the packaging of the genomic RNA into Ribonucleoprotein complexes to protect the RNA. An additional function is to enhance the stability of the entire virion through interactions with the membrane protein located in the enclosing viral membrane8. These interactions are also seen in SARS-CoV-1, where the membrane protein binds directly to the nucleocapsid via an ionic interaction1. The nucleocapsid of SARS-CoV-2 is an antagonist for interferons, suppressing the host’s defence mechanisms by preventing the synthesis of antiviral proteins9. Studies in both SARS-CoV-1 and SARS-CoV-2 have shown interactions between nucleocapsid and gRNA/sgRNA which indicate a role for the nucleocapsid in viral transcription and translation1,10. The N-Protein could also have an important role during viral assembly through interactions with envelope proteins1,11.
Many of the supplementary functions of the SARS-CoV-2 N-Proteins are still up for debate. A complete atomic structure of the RNP complex would go a long way in answering these questions, but the labile nature of the full-length N-Protein makes this a difficult task1.
5. Comparison between Coronaviruses:
The Nucleocapsid is the most conserved of the structural proteins in all coronaviruses12. This has proven useful for the development of SARS-CoV-2 Rapid Antigen Tests (for example the Roche Test). The appearance of the new English SARS-CoV-2 VUI 202012/01 variant, with changes to the spike protein, strengthens the importance of having multiple drug and test targets, particularly those that are less likely to mutate, such as the nucleocapsid13.
The high sequence analogy also allows comparison between functions within the coronavirus family, therefore a comparison between related nucleocapsids from β-Coronaviruses may shed light on these proteins’ structures and functions. The two ordered domains and the C-terminal IDR share a similar topological organization with other Coronaviruses and are involved in multiple functions in the viral life cycle. A study of the coronavirus Mouse Hepatitis Virus (MHV) analysed their nucleocapsids recruitment to Replication Transcription Complexes (RTCs) and revealed an interaction between regions on its N-terminal IDR and the serine/arginine rich region of the Linker Domain with NSP3. The interactions with NSP3 stimulate RNA replication in MHV10. Experiments have shown that the nucleocapsid from SARS-CoV-1 binds to NSP3 from MHV And interactions with NSP3 have been identified in other coronaviruses as well14,15. Thus, interactions between N-Proteins and non-structural proteins (Nsps) are proposed to have a stimulating effect for the RNA synthesis in Coronaviruses as well10.
SARS-CoV-1 and MHV both exhibit helically packed RNP complexes6 however, it is believed that SARS-CoV-2 may have a different organization16. Crystal structures of the nucleocapsid RNA binding domain from SARS-CoV-1 and SARS-CoV-2 show different crystal symmetry and packaging which could mean that SARS-CoV-2 N-Proteins have other potential contacts then SARS-CoV-14. Other possible organizations include a lattice of nucleocapsid complexes with the viral RNA linked to neighbouring RNPs like ‘beads on a string’. This ‘string‘ structure allows an efficient way of packing the large RNA genome and ensures the virus particles a high steric flexibility that is required for the incorporation into budding virions. The packaging mechanism of the SARS-CoV-2 nucleocapsid needs to be explained before we can expect to deduce an effective therapeutic approach or vaccination mechanism17.
Sources:
- 1.McBride R, van Zyl M, Fielding B. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses. Published online August 7, 2014:2991-3018. doi:10.3390/v6082991
- 2.Cubuk J, Alston JJ, Incicco JJ, et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Published online June 18, 2020. doi:10.1101/2020.06.17.158121
- 3.Peng Y, Du N, Lei Y, et al. Structures of the SARS ‐CoV‐2 nucleocapsid and their perspectives for drug design. EMBO J. Published online September 11, 2020. doi:10.15252/embj.2020105938
- 4.Kang S, Yang M, Hong Z, et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceutica Sinica B. Published online July 2020:1228-1238. doi:10.1016/j.apsb.2020.04.009
- 5.Zhou R, Zeng R, von Brunn A, Lei J. Structural characterization of the C-terminal domain of SARS-CoV-2 nucleocapsid protein. Mol Biomed. Published online August 6, 2020. doi:10.1186/s43556-020-00001-4
- 6.Chang C, Hou M-H, Chang C-F, Hsiao C-D, Huang T. The SARS coronavirus nucleocapsid protein – Forms and functions. Antiviral Research. Published online March 2014:39-50. doi:10.1016/j.antiviral.2013.12.009
- 7.Lutomski CA, El-Baba TJ, Bolla JR, Robinson CV. Proteoforms of the SARS-CoV-2 nucleocapsid protein are primed to proliferate the virus and attenuate the antibody response. Published online October 6, 2020. doi:10.1101/2020.10.06.328112
- 8.Lu S, Ye Q, Singh D, Villa E, Cleveland DW, Corbett KD. The SARS-CoV-2 Nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Published online July 31, 2020. doi:10.1101/2020.07.30.228023
- 9.Mu J, Fang Y, Yang Q, et al. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. Published online September 15, 2020. doi:10.1038/s41421-020-00208-3
- 10.Cascarina SM, Ross ED. A proposed role for the SARS‐CoV‐2 nucleocapsid protein in the formation and regulation of biomolecular condensates. FASEB j. Published online June 20, 2020:9832-9842. doi:10.1096/fj.202001351
- 11.Chen H, Cui Y, Han X, et al. Liquid–liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell Res. Published online September 8, 2020:1143-1145. doi:10.1038/s41422-020-00408-2
- 12.Chechetkin VR, Lobzin VV. Ribonucleocapsid assembly/packaging signals in the genomes of the coronaviruses SARS-CoV and SARS-CoV-2: detection, comparison and implications for therapeutic targeting. Journal of Biomolecular Structure and Dynamics. Published online September 9, 2020:1-15. doi:10.1080/07391102.2020.1815581
- 13.Diagnostics Roche. SARS-CoV-2 Rapid Antigen Test. diagnostics.roche. Published February 22, 2021. Accessed February 22, 2021. https://diagnostics.roche.com/global/en/products/params/sars-cov-2-rapid-antigen-test.html
- 14.Hurst KR, Ye R, Goebel SJ, Jayaraman P, Masters PS. An Interaction between the Nucleocapsid Protein and a Component of the Replicase-Transcriptase Complex Is Crucial for the Infectivity of Coronavirus Genomic RNA. JVI. Published online July 21, 2010:10276-10288. doi:10.1128/jvi.01287-10
- 15.Cong Y, Ulasli M, Schepers H, et al. Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle. Dutch RE, ed. J Virol. Published online November 15, 2019. doi:10.1128/jvi.01925-19
- 16.Hurst KR, Koetzner CA, Masters PS. Characterization of a Critical Interaction between the Coronavirus Nucleocapsid Protein and Nonstructural Protein 3 of the Viral Replicase-Transcriptase Complex. Journal of Virology. Published online June 12, 2013:9159-9172. doi:10.1128/jvi.01275-13
- 17.Klein S, Cortese M, Winter SL, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Published online June 23, 2020. doi:10.1101/2020.06.23.167064
Overview
During the Corona-dominated year 2020 scientists all over the world united and gathered as much information as possible to understand the exact mechanism behind the lifecycle of SARS-CoV-2.
The main question was: how can we stop the virus from invading the human cell and causing COVID-19? A focus in the quest to answer this question, was the SARS-CoV-2 entry mechanism. The group of Janet Iwasa contributes to this ongoing research process by providing a high-quality video animation of the SARS-CoV-2 entry into the human host cell. This current version of the entry animation has already been shown on PBS News (08.12.20) and we aim to improve it with your help in 2021 (see below)!
The Entry Animation
Click this Link to see the Entry Animation on YouTube.
This entry animation is a collection of current knowledge about the SARS-CoV-2 entry mechanism. What we know at this point is that the mechanism starts with the viral approach. An individual can be infected with SARS-CoV-2 after inhaling airborne viral particles. These viruses can then travel into the airways, where they may encounter host cells of the respiratory epithelium in the trachea and lungs.
As you can read in a previous blogpost, the Spikes (teal) are Corona’s key to invade the host cell and thus of great interest in terms of vaccination and therapeutic approaches against COVID-19. The Spike protein recognizes a specific receptor on the human host cell surface, called ACE2 (purple). Usually, the Spikes are very dynamic and able to undergo opening, closing and bending movements. But after binding to ACE2, the protein is locked into its open position. Another protein on the cell surface, called TMPRSS2 (orange), can then come along and cut the Spike protein in a specific location. These segments of the Spike protein fall away, exposing a portion of the Spike protein which was previously hidden.
The Spike protein is then able to undergo a series of dramatic conformational changes. During the first stage, the Spike protein inserts itself into the membrane of the cell. In the second stage, segments of the Spike protein zipper back on itself, forcing the membrane of the cell and the viral membrane to fuse. After fusion, the viral RNA is deposited into the host cell, where it will direct the cell to produce more virions. This process is known as post-fusion.
The Annotation Tool
Click this Link to use the Annotation Tool.
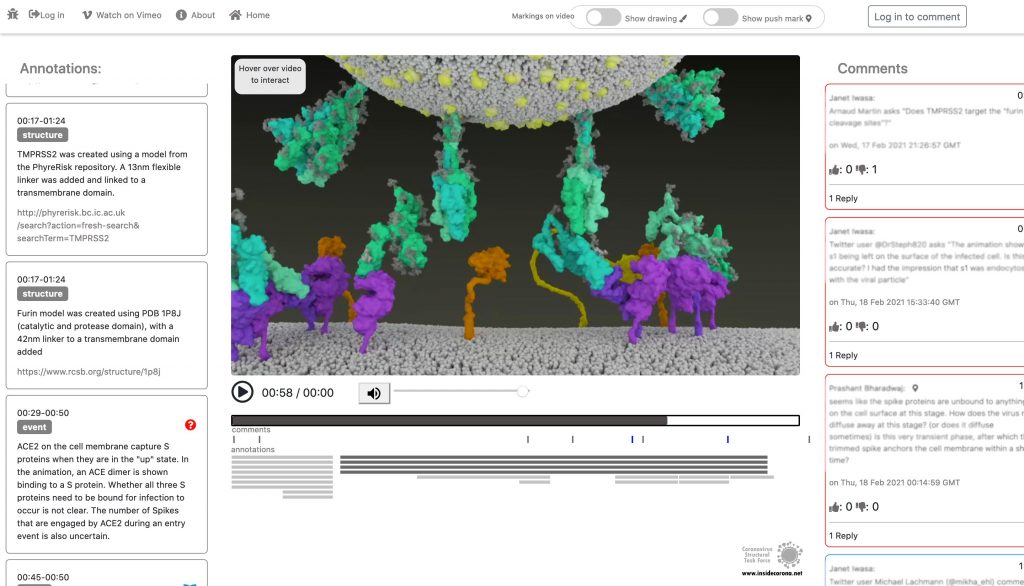
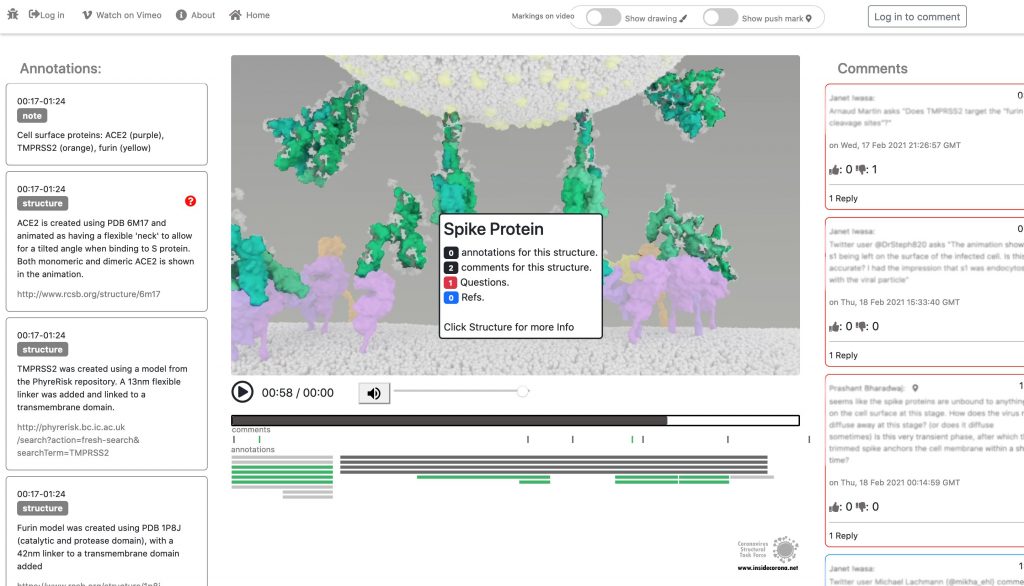
In January, this will be supplemented with a tool so that the knowledge about the SARS-CoV-2 entry mechanism can be discussed interactively by scientists all over the world. This online platform will serve as a basis for scientific discussion by providing an annotation tool. Scientific users can set a pin at any point of the video and comment their suggestions, criticism or questions about the mechanism and the structure depictions (see Fig. 1 for a prototype). Based on these annotations, the Iwasa Group will improve the animation of the entry process to provide an up-to-date detailed representation of this key process. The resulting entry animation is not only addressed to scientists, but it is also used for public outreach and education.
Even though the entry mechanism is not entirely understood yet, it could already be depicted in the fantastic animation of the Iwasa Group. There are still a lot of details and additional information to be found out about this process. From January on, the annotation tool therefore will provide the opportunity to discuss this mechanism publicly.
Thanks to the Iwasa Group for this Christmas present!
Merry Christmas!
Introduction
It is known as VUI‑202012/01 or B.1.1.7 – the new mutation of the coronavirus Sars-CoV-2. It may be responsible for a sharply increased number of infections in the southeast of England (1), however, the scientific results leading to very strict lockdown measurements in the south of the UK, and travel restrictions across Europe are few and far between. Here, we have compiled what is known up until now.
On mutations
Mutations are normal in the evolution of life – and of viruses. If two similar viruses have infected the same cell, their genomes can become mixed-up, one of the reasons why animal influenza strains are considered so dangerous. This is also called recombination. Mutations can be caused by chemicals, radiation (including UV light) and errors during genome copying. A typical SARS-CoV-2 virus accumulates two amino acid changes per month in its genome — a rate of change about half that of influenza (2). This is because SARS-CoV-2 can repair RNA to some extent. But even so, this natural process led to thousands of mutations since the beginning of the pandemic. If they affected the virus life cycle negatively, that strain may have likely died out - if they did not make a difference or enhanced its chances of survival, it may have persisted.
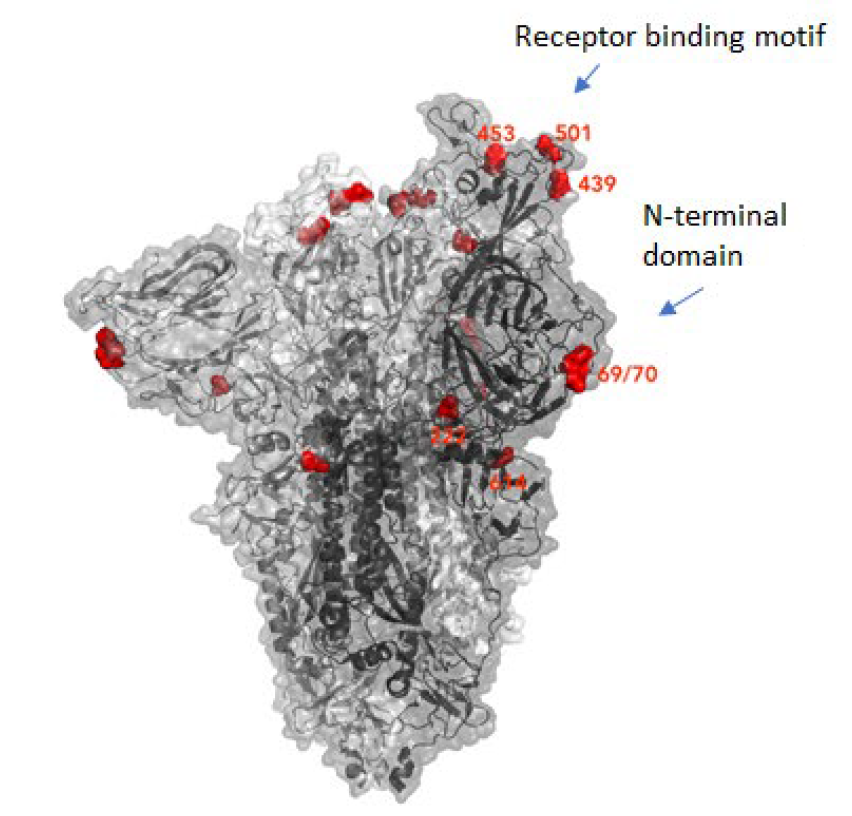
Many mutations that are observed occur in the spike protein, which both serves to recognize potential host cells but is also what is being recognized by antibodies (i.e., the immune system).
Changes here can be crucial for the survival of the virus (“evolutionary pressure”) as they could significantly alter its affinity to the human receptor ACE2, which the virus uses as gateway to our cells.
What vaccines do
Most, if not all, potential COVID-19 vaccines expose our body to some part of the spike protein, which can be made by the body itself (mRNA vaccines) or carried by a harmless virus instead of SARS-CoV-2 (vector). Our body then produces antibodies which specifically recognize the spike and persist for several months. If we are exposed afterwards to the real virus, the body can recognize it immediately – and the risk of infection is much lower as the immune system swings into action immediately. Earlier this year, the spike mutation D614G (amino acid residue number 614 changing from aspartic acid (D) to glycine (G)) caused quite a stir in the media, and became the predominant form of SARS-CoV-2 (2, 3). However, if and in how far this was caused by natural selection is still debated (3). Another example which triggered an increased media coverage was the mutation Spike Y453F, which originated from infected minks in Denmark (4) and led to a culling of millions of animals. In any case, if we would be vaccinated with a spike protein form that would be different from the one in a virus we encounter later, there is a small chance that the vaccine may be rendered ineffective. This chance is, however, small for SARS-CoV-2, in any case much smaller than for HIV, which famously evaded any attempt to develop a vaccine.

What do we know?
There was a steep rise in infections in the UK recently, as in most other European countries.
A new mutation of the virus has emerged and seems to replace the old version of SARS-CoV-2 (5). Thousands of patients have been found to carry this variant.
This new variant has more mutations at once than expected. These mutations have not observed in this combination before.
The variant has been reported in the UK, the Netherlands, Denmark, Australia and Belgium so far.
What is striking to me as scientist about these findings is one thing in particular: How could the British government find that thousands of people were having the new SARS-CoV-2 variant, instead of the old, if the illness does not look any different? Sequencing samples from each and every patient would be technically very challenging, if not impossible. How could they know? The answer is:
Serendipity
The main PCR test employed in the United Kingdom is Thermo Fisher's TaqPathCOVID-19. This test identifies RNA on three different genome locations: In ORF1ab, nucleotide and spike. Now, it stopped working for the spike portion of the test, while the other two RNAs were still found to be present, which likely prompted scientists to sequence some of the samples in question. And indeed, the new mutant has a deletion of histidine-69 and valine-70, called 69-70del. This permitted easy differentiation of patients with the old SARS-CoV-2 (3 hits) and the new (2 hits) and is the reason why we know so much about the epidemiology of this variant!* It has also to be said that this test is not used as often in other countries, such as Germany, and this could well be the reason why we do not know if and how widespread it is here. In addition, other countries sequence much smaller proportions of virus isolates than the UK, so ongoing circulation of this variant outside of the UK cannot be excluded.
The details of the mutation
The new variant of SARS-CoV-2 VUI-202012/01 has 14 amino acid changes and three deletions affecting the genes for ORF1ab, spike and ORF8. One of these mutations (N501Y) occurs in the receptor binding domain and could lead to an increased binding affinity to the human ACE2. The 69-70 deletion has likely an immunological role and is the reason this mutant was detected so widely, as this RNA location is used for PCR tests. Another interesting mutation is the P681H, which is next to a furin cleavage site that has a biological significance in membrane fusion. These mutations could be responsible for the increased transmissibility. The effects of the other mutations aren’t fully investigated yet. Here is a list of the mutations which have been observed in the VUI‑202012/01 or B.1.1.7 variant:
| T1001I in gene ORF1ab | |
| A1708D in gene ORF1ab | |
| I2230T in gene ORF1ab | |
| SGF 3675-3677 deletion in gene ORF1ab | |
| A1708D in gene ORF1ab | |
| HV 69-70 deletion in spike | The 69-70 deletion on the spike protein is a re-occurring mutation that has shown to often co-occur with other amino acid changes in the RBD (6, 7). (1) Evasion to the human immune response and in association with other receptor binding domain changes (1) (2) Immunological role (8) (3) Leads to diagnostic failures which permit detection (see above, "Serendipity") (4) Associated with immune escape in immunocompromised patients (9(8)) Furthermore, the 69-70 deletion arose in multiple unrelated lineages and is associated with the evasion of the immune response (9). It is being hypothesized that this mutation undergoes a strong positive selection when exposed to convalescent plasma therapy in an immunocompromised human host (7). |
| Y144 deletion in spike | Deletion in the spike N-terminal domain (9) |
| N501Y in spike | One of six key contact residues in the spike receptor binding domains, this mutation leads to an increasing binding affinity to human and murine ACE2 (1). |
| A570D in spike | Mutation located at the spike receptor binding domain (10) |
| P681H in spike | The P681H mutation is located directly next to the furin cleavage site. It is one of the four residues which are insertions when compared to closely related coronaviruses, creating a furin cleavage site in the spike protein between the spike S1 and S2 domains. This prompts the entry of the virus into respiratory epithelial cells as well as the transmission in animal models (1) The S1/S2 furin cleavage site of SARS-CoV-2 is not found in closely related coronaviruses and has been shown to promote entry into respiratory epithelial cells and transmission in animal models (9) |
| T716I in spike | Mutation in in the S2 domain |
| S982A in spike | Mutation in in the S2 domain (10) |
| D1118H in spike | Mutation in in the S2 domain (8) |
| Q27 stop in ORF8 | The Q27stop mutation in the ORF8 leads to the truncation of the ORF8, and as it only consists of 121 amino acids, the consequence might be a loss of function. These and the other mutations could be responsible for the increased transmissibility of the B.1.1.7 variant. In any case, this mutation truncates the ORF8 protein at residue 27 or renders it inactive which allows further downstream mutations to accrue. (1) |
| R52I in ORF8 | |
| Y73C in ORF8 | |
| D3L in nucleocapsid | |
| S235F in nucleocapsid |
Why were there so many mutations at once?
This could be a result of prolonged or chronical SARS-CoV-2 infections as study of these infections reveal unusually large numbers of nucleotide changes and deletion mutations and often high ratios of non-synonymous changes. In addition to this, convalescent plasma treatment can cause intra-patient virus genetic diversity (11).
What does the new mutation mean in terms of impact and epidemiology?
There was an increase in cases with the new strain in total and in
proportion to the old (1). What does that mean for us?
This is what the internet says:
The COVID-19 genomics UK consortium (COG) reports about a “priority set of SARS-CoV-2 Spike mutations that are of particular interest based on potential epidemiological significance in the UK and/or biological evidence based on the literature or unpublished work.” (9)
The New and Emerging Respiratory Virus Threats Advisory Group of the British government (NERVTAG) discussed the new variant on Friday and concluded that its growth rate is higher by 67-75% and that this is likely due to a selective advantage. “In summary, NERVTAG has moderate confidence that VUI-202012/01 demonstrates a substantial increase in transmissibility compared to other variants.” (12) This is very likely the source of Boris Johnson’s claim to this strain being “70% more infectious”.
The English government writes that PHE (Public Health England) „is working with partners to investigate and plans to share its findings over the next 2 weeks. There is currently no evidence to suggest that the variant has any impact on disease severity, antibody response or vaccine efficacy. High numbers of cases of the variant virus have been observed in some areas where there is also a high incidence of COVID-19. It is not yet known whether the variant is responsible for these increased numbers of cases.” (13)
Conclusion
From this, we conclude that the British government, and we, do not know yet. It has not been conclusively shown that the new variant is more infectious (likely), has an easier time to evade the host immune system or if the vaccine will be less effective against it (very unlikely). The epidemologic model which predicts a higher tranmissability has still to be published, the science is still in the making. Tests of vaccines against the new variant are ongoing and will take a few weeks. There is yet little evidence that this new variant poses a significantly bigger threat than others - or to the contrary.
Acknowledgements
While I am listed as author of this article, it could not have been written without the help and research by Pairoh Seeliger, Lea von Soosten, Luise Kandler, Erik Nebelung and Oliver Kippes who all helped in this.
I would also thank Nicolai Wilk from Thermo Fisher Scientific who quickly responded to my questions about their test.
The title picture shows mutation cards from the game Pandemic Expansion: On the Brink by Z-Man Games.
- *The 69-70del mutation is predominantly observed in B.1.1 (including B.1.1.7), B.1.258, and the cluster 5 variant lineages of SARS-CoV-2.
References
- 1.A. Rambaut, Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. virological.org (2020), (available at https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563).
- 2.E. Callaway, The coronavirus is mutating — does it matter? Nature, 174–177 (2020).
- 3.L. Zhang, C. B. Jackson, H. Mou, A. Ojha, H. Peng, B. D. Quinlan, E. S. Rangarajan, A. Pan, A. Vanderheiden, M. S. Suthar, W. Li, T. Izard, C. Rader, M. Farzan, H. Choe, SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun (2020), doi:10.1038/s41467-020-19808-4.
- 4.ECDC, Detection of new SARS-CoV-2 variants related to mink. www.ecdc.europa.eu (2020), (available at https://www.ecdc.europa.eu/sites/default/files/documents/RRA-SARS-CoV-2-in-mink-12-nov-2020.pdf).
- 5.ONS UK , Percentage of COVID-19 cases that are positive for ORF1ab and N genes. www.ons.gov.uk (2020), (available at https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/adhocs/12690percentageofcovid19casesthatarepositivefororf1abandngenes).
- 6.R. M. Dawood, M. A. El-Meguid, G. M. Salum, K. El-Wakeel, M. Shemis, M. K. El Awady, Bioinformatics prediction of B and T cell epitopes within the spike and nucleocapsid proteins of SARS-CoV2. Journal of Infection and Public Health (2020), doi:10.1016/j.jiph.2020.12.006.
- 7.S. A. Kemp, D. A. Collier, R. Datir, S. Gayed, A. Jahun, M. Hosmillo, I. A. Ferreira, C. Rees-Spear, P. Mlcochova, I. U. Lumb, D. Roberts, A. Chandra, N. Temperton, K. Sharrocks, E. Blane, J. A. Briggs, K. G. Smith, J. R. Bradley, C. Smith, R. Goldstein, I. G. Goodfellow, A. Smielewska, J. P. Skittrall, T. Gouliouris, E. Gkrania-Klotsas, C. J. Illingworth, L. E. McCoy, R. K. Gupta, Neutralising antibodies drive Spike mediated SARS-CoV-2 evasion (2020), , doi:10.1101/2020.12.05.20241927.
- 8.K. Kupferschmidt, Mutant coronavirus in the United Kingdom sets off alarms, but its importance remains unclear. Science (2020), doi:10.1126/science.abg2626.
- 9.COG, COG-UK update on SARS-CoV-2 Spike mutations of special interest Report 1. https://www.cogconsortium.uk (2020), (available at https://www.cogconsortium.uk/wp-content/uploads/2020/12/Report-1_COG-UK_19-December-2020_SARS-CoV-2-Mutations.pdf).
- 10.S. Kemp, W. Harvey, R. Datir, D. Collier, I. Ferreira, A. Carabelii, D. L. Robertson, R. K. Gupta, Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion ΔH69/V70 (2020), , doi:10.1101/2020.12.14.422555.
- 11.ECDC, Threat Assessment Brief: Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. www.ecdc.europa.eu (2020), (available at https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-rapid-increase-sars-cov-2-variant-united-kingdom).
- 12.NERVTAG, NERVTAG meeting on SARS-CoV-2 variant under investigation VUI-202012/01. https://khub.net (2020), (available at https://khub.net/documents/135939561/338928724/SARS-CoV-2+variant+under+investigation%2C+meeting+minutes.pdf/962e866b-161f-2fd5-1030-32b6ab467896?t=1608470511452).
- 13.PHE, PHE investigating a novel variant of COVID-19 . www.gov.uk (2020), (available at https://www.gov.uk/government/news/phe-investigating-a-novel-variant-of-covid-19).
On Nov 9th, 2020 Pfizer issued a press release stating their conclusion that the COVID-19 vaccine they developed with BioNTech appeared to be 90% effective. While their test contained over 43,000 volunteers they had only detected 94 cases of COVID-19. How confident can you be with only 94 cases? I decided to explore this matter for myself.
I am but a lowly crystallographer, and I’m sure a proper mathematician could do a more rigorous job, but I’ll do the best I can.
The Experimental Design
I was not familiar with the design of this clinical trial but it seems rather straight-forward. You take a whole lot of people and split them into two groups, keeping their group assignment secret from everyone who will be involved in their handling until the end of the trial. The members of one group are given the treatment which we hope is a vaccine while the others are given a sham treatment which is indistinguishable from the “vaccine” by both the participants and their doctors. You then wait to see if anybody comes down with COVID-19.
How long do you wait? You want to wait until you have enough cases to reliably answer the question you hope the study will answer but, to avoid bias, the end point has to be set before the start. If you constantly watch the results and decide to stop when the numbers look good, you could claim success when there is none. After all, life is filled with statistical fluctuations and the results might get worst with longer time.
The press release says that the design of this test says to end after 164 cases of COVID-19 arose among the volunteers but they would peek at the results after 32, 62, and 94 cases. For unspecified reasons they skipped the peek at 32 and, it appears, that the case count shot up to the 94 case trigger while they were discussing the merit of the 62 threshold. I guess this is the only benefit to the world of the huge surge in COVID-19 cases this fall.
It was the 94 case checkpoint that led them to conclude that it was likely that their vaccine candidate was 90% effective at preventing the disease.
But how likely?
To judge the reliability of the 90% number I’ll need to do some statistics. That “proper mathematician” I mentioned earlier would be able to pull out the expected distributions for the experimental results and precisely calculate probabilities and likelihoods. That knowledge is not in my skill set so I’m left with running simulations.
I wrote a program in Mathematica Script to generate many simulations of vaccine trials and then examined their variability. This program has a loop that produces a person with a 100% chance of developing COVID-19 without intervention. That person is assigned either to the Placebo or Vaccinated group. Those poor souls put in the Placebo group are counted as COVID-19 cases. Those in the Vaccinated group are only sickened if they lose a roll of the dice. For each series of simulations I assume a level of effectively for the vaccine. If the run is for a vaccine with 30% effectiveness, for example, the volunteer only get sick if they roll over 30 (okay, I’m using percentile dice.) Those folk protected by the vaccine are let go and the sick are counted as vaccine failures. When the total number of sick reaches the target that trial is complete, the number of sick in each group recorded, and the next trial is started. To ensure that I have a good sample of all possible clinical trials, I simulated a hundred thousand trials for each assumed efficacy.
To keep the numbers simple I ran 100 case trials instead of 94.
When the vaccine is ineffective (can we still call such a thing a vaccine?) there will be an equal number of COVID-19 cases in the Placebo and Vaccinated groups, and this number will be around 50 but there will be variation. If the vaccine is 100% effective the vaccinated group will be completely protected and all 100 cases will be in the Placebo group. The key result of a vaccine trial is the difference in the number of cases. This difference can never be greater than 100 because there aren’t enough cases to result in a bigger number. The difference can, however, be negative since it is possible to have more cases in the Vaccinated group. The most likely explanation for such a result is that the vaccine is very ineffective and randomness of infections happens to result this odd distribution.
Here are my results for a series of hypothetical vaccines with varying efficacy.
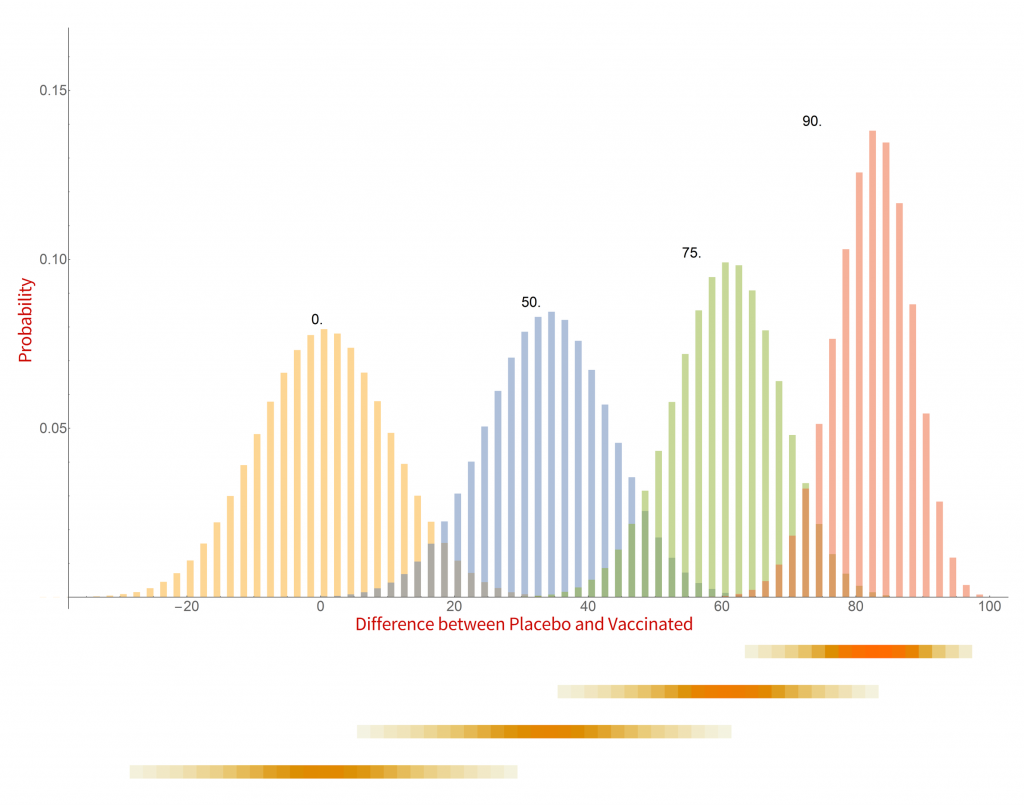
Histograms of the probability of a clinical trial of a vaccine with an assumed efficacy resulting in a particular difference in COVID-19 case numbers between the placebo and vaccinated groups. CC-BY-NC Dale E. Tronrud / Coronavirus Structural Task Force
There are a whole lot of interesting things in this graph. When the vaccine is completely ineffective the most common result of a trial is a difference of zero between the Placebo and Vaccinated groups. There is a fairly wide distribution of results that occur, however. This is the result of statistical fluctuations due to the small number of cases of COVID-19 in the sample (here 100). The distributions for all the simulated efficacies have about the same width, with the exception of those near 100%. Since the difference can never be larger than 100 those distributions get sharper and develop a tail on the lower side.
Let’s look at some scenarios. The graph shows that the most common result of a trial of a vaccine with efficacy of 50% has a difference in number of cases between the Placebo and Vaccinated groups of 36, but sometimes the difference is larger and sometimes smaller. If the vaccine was worthless the most common trial result is zero, but there is also variability. The two histograms overlap considerably which indicates that one cannot distinguish between an efficacy of zero or 50% if the difference in the number of cases in your trial is in the range of zero to about 36. If the difference is greater than this you could conclude that the vaccine is more likely to be 50% than zero percent, and zero percent is the more likely of the two if the difference is negative. Still, there is a wide range of possible outcomes of a trial that have ambiguous interpretation.
On the other hand, what if we have a difference of 80 (90 cases in the Placebo group and 10 in the Vaccinated group)? There isn’t any significant overlap between zero percent efficacy and 90% at the point where the difference is 80. It is much more likely that the vaccine is 90% effective than zero. There is overlap with the 75% effectiveness histogram and we have to admit that it is possible that the vaccine is only 75% effective, but 90% is more likely.
This leads us to realize that the result of a vaccine trial has to result in a range of possible efficacies, with a varying probability of each. My little plot doesn’t make such an assessment very easy. In fact, the plot is starting to show some problems. What it shows is the probability of a trial having a particular result given the effectiveness of the vaccine. What we really want is the probability of each possible efficacy given the result of the clinical trial.
We have to transform our probabilities!
Turning everything on its head
While the calculations I just discussed were easy to set up and understand, they do not really reflect the experiment being done in a vaccine trial. I was assuming an effectiveness of the vaccine and running many, many trials. In reality the effectiveness is unknown and only one trial is run. Where I calculated the probability of a particular difference in COVID-19 cases given the effectiveness of the vaccine what I really want is the probability of the effectiveness of the vaccine given the results of a single clinical trial. It is often difficult to devise such a calculation from scratch but it is pretty straight forward to calculate it from the results I already have.
The first step toward the proper calculation is to expand the current plot. My first figure included simulations of just four possible efficacies. To display more possibilities, I need to abandon histograms. At the bottom of the plot I show four color-shaded bars. In these bars the color is darker when the corresponding histogram is taller. While not as visually clear these bars have the advantage that they can be stacked, and many more plotted in a single figure.
With this new tool I can calculate and display the simulated distribution of clinical trials for every vaccine efficacy from 0% to 100% in 1% steps. The new chart is displayed here.
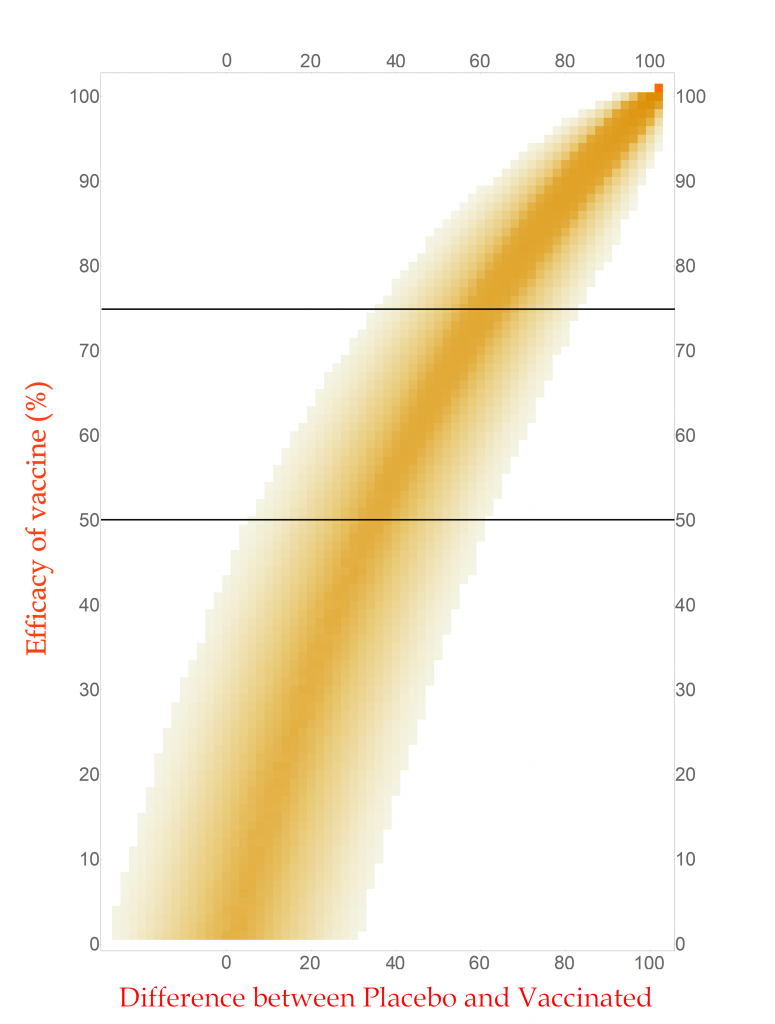
CC-BY-NC Dale E. Tronrud / Coronavirus Structural Task Force
This chart is read by locating the efficacy of your vaccine on the vertical axis and drawing a horizontal line there. The pattern of colors along that line represents the probability of each difference in cases between the Placebo and Vaccinated groups in a clinical trial. I have drawn two such lines, one for a vaccine with 75% effectiveness and another for one with 50%. You can see that the most likely result for the 75% one is about 60 cases (20 in Vaccinated and 80 in Placebo) and the other at about 34 cases. (You figure it out.) In this plot you can see the continuous change as the efficacy of the vaccine is changed. The key point is that there is a spread of results, but I described that before.
With any set of probabilities the full set has to always add up to one. For each horizontal line of colored boxes in this plot the sum of their probabilities is one. A set of numbers with this property is said to be “normalized”.
The vertical lines are not normalized in this plot, as you can see by looking at its left side. There are just a few, very lightly colored or low probability boxes and above them is simply white, which represents zero probability (or at least very, very, very small). This side of the plot has a difference in COVID-19 cases of -24, or in other words the Vaccinated group had 24 more cases of disease than the Placebo. Such an outcome for a clinical trial is very unlikely for any vaccine that has even a tiny amount of success (and is pretty unlikely for one that is merely useless).
Since the probabilities along vertical lines are not normalized they cannot be used as a histogram. Conveniently for us, this can be corrected simply by normalizing them. This is done by summing all the probabilities along each vertical line in this plot and dividing the probabilities in the line by that sum. This gives us a new set of probabilities and a new plot.
How does this magic work? The procedure is justified by a hundreds-of-years-old mathematical theorem called Bayes’s Law. This blog post is already getting long and I leave the application of your favorite search engine to you.
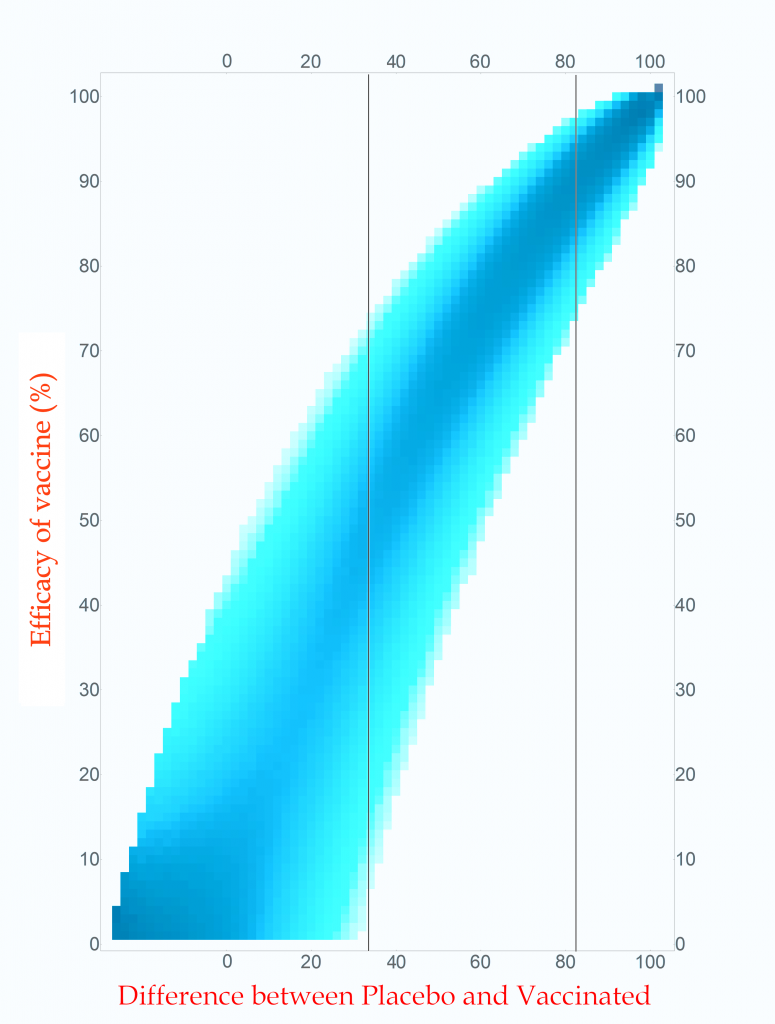
Probability of vaccine effectiveness as a function of clinical trial outcome.
CC-BY-NC Dale E. Tronrud / Coronavirus Structural Task Force
The first thing to note is that the new plot isn’t much different than the original. While the lower-left side has clearly changed, that area is not very interesting. On the right, where the action is, it looks the same. For this reason, many fields of science simply use the unnormalized plot.
The new plot allows us to draw vertical lines (but forbids horizontal lines!). I have drawn example lines at differences of 36 cases and 82 cases. If our clinical trial results in a difference in cases of 82 (91 in Placebo and 9 in Vaccinated) we can see from the line on the plot that the most probable effectiveness of that vaccine is about 90%! This is very close to the happy number reported in Pfizer’s press release. The plot also shows us that there is uncertainty in this number. The vaccine’s effectiveness could be in the mid 70’s or in the upper 90’s.
This is the nature of all experimental work. All results have uncertainties and it is as important to know the amount of uncertainty as it is to know the direct result. You can see how important this is by looking at the 36 case difference line. The darkest blocks along this line, and therefore the most probable efficacy, are near 50%. This would also indicate a useful vaccine, but look at the spread! The width of the uncertainty goes all the way to zero – This vaccine could be worthless. A clinical trial that waited until only 100 cases occurred cannot distinguish between a vaccine with 50% efficacy and a worthless one.
If you wait for more cases to develop the width of the stripe in the plot becomes narrower and the uncertainty drops. The goal of Pfizer was to develop a vaccine of at least 50% efficacy so their design was to wait for 164 cases to give them a narrow enough band to clearly distinguish 50% from zero percent. Just in case the vaccine was better than 50% they built into the design of their trial several points where they could peek and see what was going on. They, and we, lucked out!
What are all those other people for?
The surprising thing about this analysis is that the total number of people in the trial is unimportant when calculating the uncertainty of the result. That answer is the same for a trial with 500 volunteers and a trial with 50,000 volunteers. The only thing that is important is the number of cases of COVID-19.
All those tens of thousands of people are important for other reasons. Very relevant to the current pandemic is that a larger number of volunteers will accumulate the target number of cases sooner: if you double the number of volunteers you will reach the target in half the time. We all want to know as quickly as possible if these vaccines are effective so we want the trials to consist of as many people as the companies can manage.
The other use for large numbers is the search for grim and hopefully rare side effects. These are likely to arise at much lower rates than viral infections and much larger numbers of volunteers are required to achieve statistical significance. A side effect that only occurs in 1 of 5,000 people will require a very large number of participants to be detected. A compounding factor is that a search for an unknown result requires many more data points than the search for a specific outcome, such as COVID-19 infection. (The checks for side effects are only now being described to the public, and I’ll not go into them here.) While the doctors keep an eye out for life threatening problems during the trial, the secret books identifying which volunteers are in which group are kept closed until the end.
End game
After all this interesting math I find that, yes, 94 diseased people are quite enough to conclude that a vaccine is effective at around a 90% level, or at least 70%.
Postscript
During the writing of this post Moderna issued a press release about their vaccine candidate. The analysis presented here applies equally to their trial since I made no assumptions at all about the nature of the vaccine being tested. The only complaint I have with their press release is that they quoted the effectiveness of their vaccine as 94.5%. As you now know, this level of precision is ridiculous. It would be better just to say it is “somewhere around 95% effective”.
This article has been written by Cameron Fyfe and Lea von Soosten.
In the previous two articles we spoke of proteins involved in RNA synthesis and proteins involved in removing errors during that process. There are also proteins produced by SARS-CoV-2 that can mimic functions of the host cell to avoid its defense mechanisms.
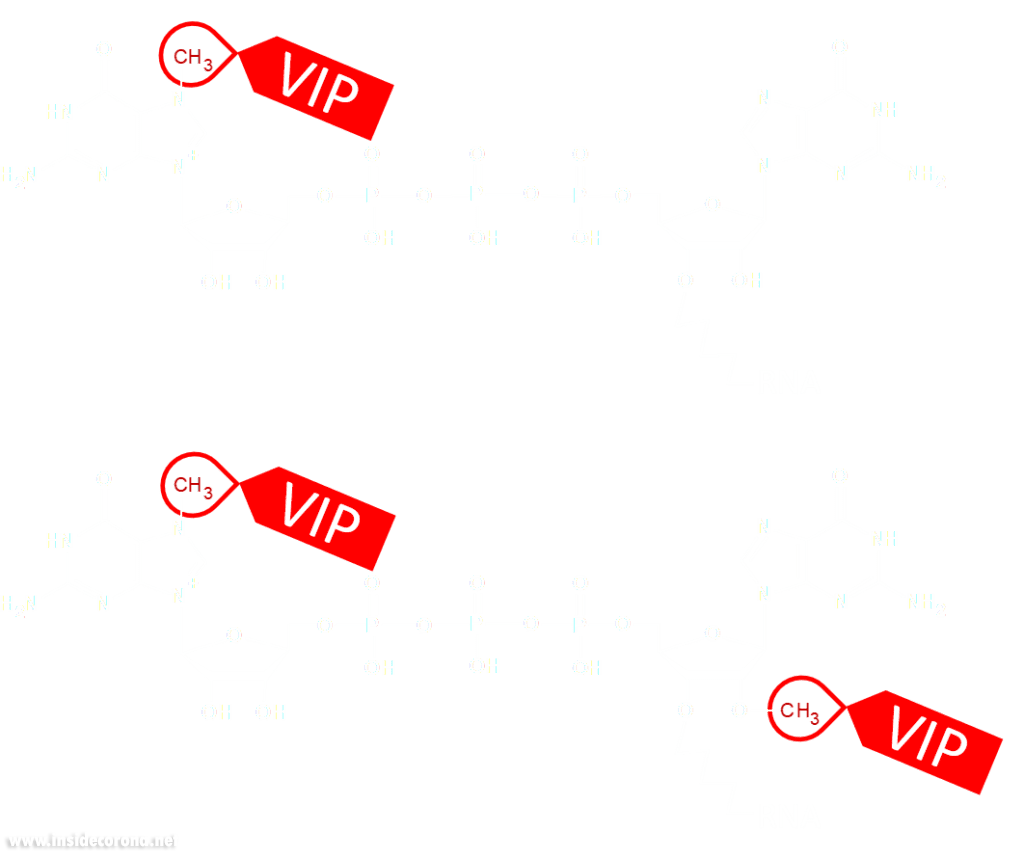
Eukaryotic cells have evolved to have various immune responses to fight infection or invasion from pathogens. One of these is to recognize and chop up any RNA that is from other organisms using enzymes called exoribonucleases. In order to differentiate "friendly" RNA from "foe" RNA is to give the cell's own RNA a VIP badge so that only unfriendly RNA will be shredded. These "VIP badges" are made of a 5’ to 5’ triphosphate linkage with two methylation modifications (see Fig. 1). In order to evade exoribonucleases, the virus SARS-CoV-2 has a way of 5’ to 5’ capping as well as adding its own methyl group VIP badges to protect its RNA from the defense mechanisms of invaded cells. Two Very Important Proteins, nsp14 and nsp16, have this methyltransferase activity using an S-Adenosyl methionine (SAM) as cofactor.
What are SAM methyltransferases?
Methyltransferase enzymes are a large superfamily of proteins that perform the chemical addition of a methyl group (a carbon with three hydrogens) to a variety of substrates. These substrates include small molecules, other proteins, DNA, and RNA 2,3. This superfamily of proteins often uses a small molecule, S-Adenosyl methionine (SAM), to transfer a methyl group to its target substrate (Figure 2). During this process, the methyl group bound to the charged sulfur is brought in proximity to the target atom of the substrate, transferring the methyl group (Figure 2), resulting in the methylated product and the byproduct S-Adenosyl homocysteine (SAH).
Methyltransferases of SARS-CoV-2
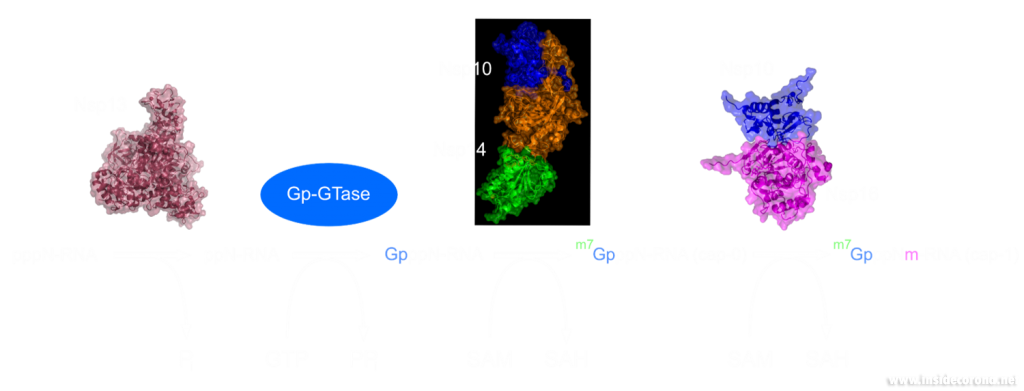
In a previous article we spoke of the exoribonuclease (ExoN) proofreading activity of Nsp14 (not to be confused with the host cell's own exoribonucleases that are part of the immune system, see above). After the 5’ to 5’ guanine triphosphate addition has been performed on the mRNA the guanine-N7-methyltransferase activity of Nsp14 comes into play producing the first Cap0 structure with a VIP tag (Figure 1, 3). Only after this methylation has been performed can Nsp16 have action and perform the second 2’O-methylation to produce the Cap1 structure (Figure 1, 3).
Not only do both of these proteins perform VIP methylations of mRNA, but they also both bind another non-structural protein, Nsp10. The binding of Nsp10 has been shown to increase activity in both Nsp14 ExoN activity and Nsp16 methyltransferase activity4. Independently, Nsp10 has also been shown to have the ability to bind both single and double stranded DNA and RNA5.
Structures of nsp14 and nsp16
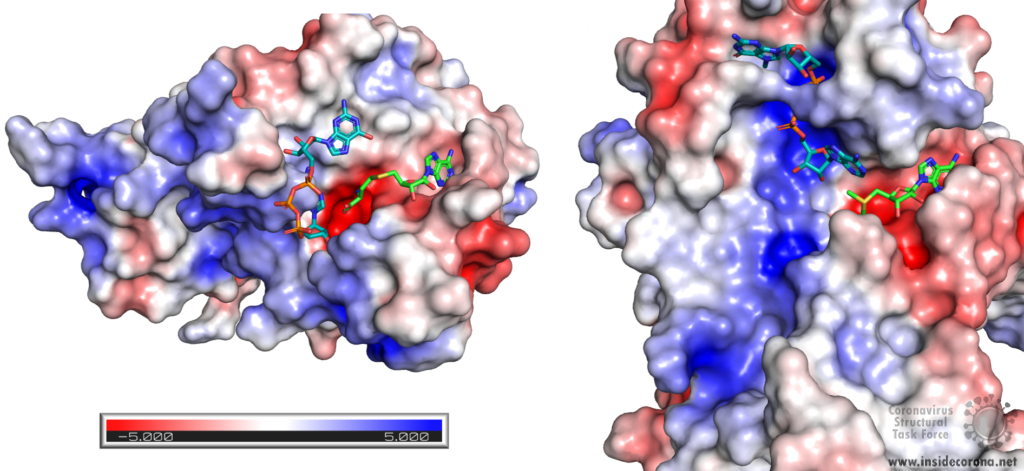
Nsp14 consists of two domains, each carrying out one specific task: the first is responsible for the ExoN activity, whilst the second executes the first methylation of the Guanosine-N7 of the RNA end cap. The two domains are connected by a flexible region that acts like a hinge, allowing movement between the domains. The second domain has an unusual and unique structure which does not follow the typical Rossmann fold seen in other SAM methyltransferases. The methyltransferase active site has a negatively charged binding pocket that holds SAM (SAH in Figure 4. A) in close proximity to the Guanosine-P3-adenosine-5',5'-triphosphate (GpppA) substrate (Figure 4A). The binding pocket holding the GpppA has a positive charge and the surface charge of the region below is also positively charged (Figure 4A). The distance between the N7 of the 5’ Guanosine and the sulfur that transfers the methyl group is 4.4 Å5,6. This close proximity of cofactor and substrate facilitates the methylation.
Similar to Nsp14, Nsp16 has a negatively charged binding pocket to position SAM in close proximity to the m7GpppA substrate (Figure 4. B). The m7GpppA binding site has a positive charge. The space nearby the 3’ end of the m7GpppA also has an overall positive charge and would be expected to bind the extension of the full length RNA (Figure 4. B)4. The distance between the methyl group and the sulfur of SAM and the 2’O of the m7GpppA substrate is 3.1Å and 4.9Å, respectively.
Structure of nsp10 and its function
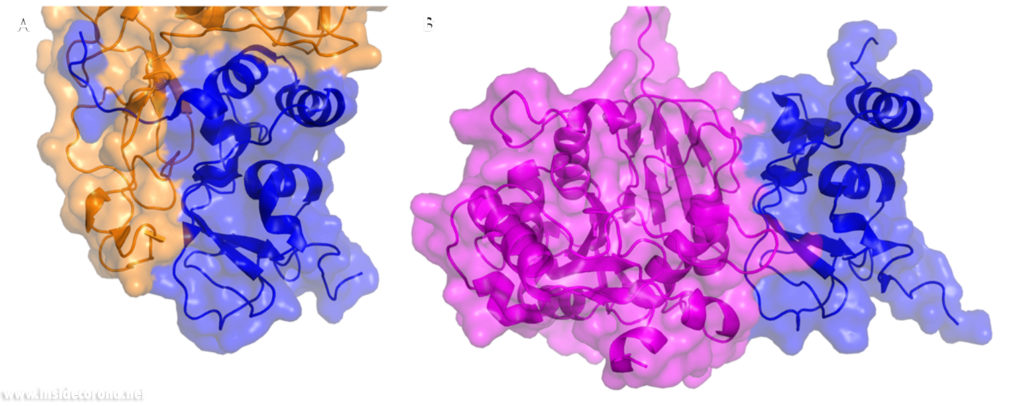
In a previous article where we spoke about the exoribonuclease (ExoN) activity of the first domain of nsp14, we highlighted the interaction between nsp14 and nsp10 (Figure 5A). This is quite significant, as the activity of ExoN increases 30-fold when nsp10 and nsp14 are bound. Nsp10 also functions as a co-factor for nsp16, stabilizing the SAM-binding pocket7 and enhances its methyltransferase enzymatic activity significantly4 (Figure 5B). For SARS-CoV, and similarly for MERS-CoV, the affinity for m7GpppA-RNA and m7GpppA cap analogue of nsp16 was found to be low until binding to nsp10, which enhanced the affinity for binding to RNA8,9. With a reduced activity in Nsp16 in the absence of Nsp10 and a huge decrease in activity of the exonuclease domain of Nsp14, interfering with these interactions could result in decreased viability of COVID-19.
Methyltransferases Nsp14 and Nsp16 as drugs targets
As both Nsp14 and Nsp16 use the cofactor SAM and have affinity for the endcap of RNA, these two binding sites could be worthwhile targets for drug development in the fight against SARS-CoV-2. Without the VIP status provided by the methylation of RNA the host immune system could defend against the viral RNA. It might be possible to block these binding pockets by letting the protein bind to something that is similar to SAM, which cannot function as a methyl donor. An additional challenge is that the inhibitor has to be very specific to Nsp14 or Nsp16, so as not to affect similar human proteins in a negative way.
Sinefungin is a 5’-aminoalkyl analog of SAH and SAM, which can do exactly that: it has the ability to inhibit all SAM methyltransferases (Figure 6). Sinefungin was first discovered in 1973 from Strepromyces griseolus and was described as having antifungal antibiotic properties10.
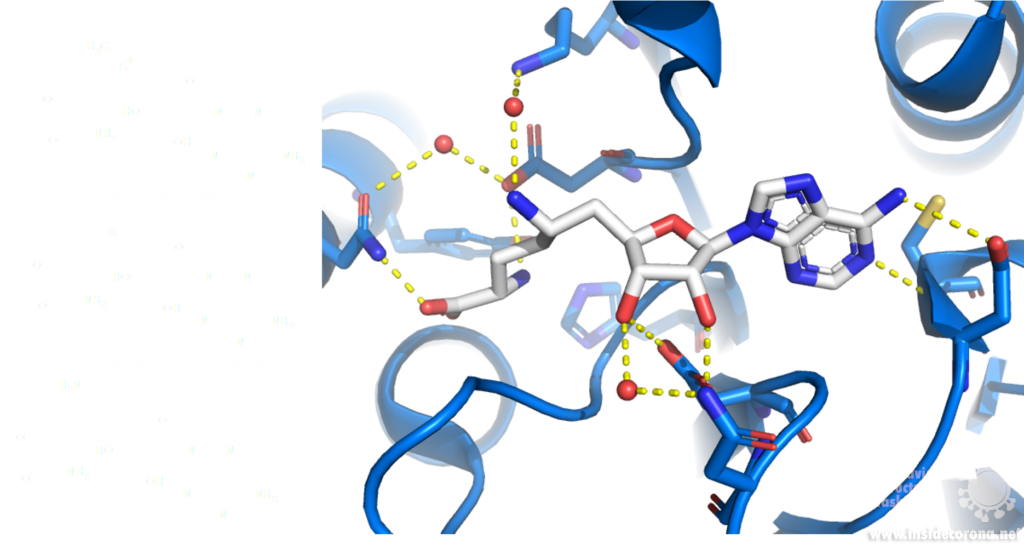
A major issue with targeting the SAM binding site of Nsps with compounds such as sinefungin (Figure 6) is that there are many proteins within humans that use SAM as a cofactor for normal function. This results in singefungin and other similar compounds having toxic effects on human cells. Synthetic chemists have already been able to synthesize analogs of sinefungin with improved affinities to specific SAM methyltransferases. Recently, specific inhibitors have been developed to target a nicotinamide SAM methyltransferase11. This inhibitor was developed to have affinity to both the cofactor binding site and the substrate binding site by combining the nicotinamide substrate with the SAM cofactor. Recent work has looked at how singefungin binds to the active site of Nsp16 in order to have a detailed understanding of its interaction to design more specific inhibitors that can target methyltransferases from SARS-CoV-24. Similar to the development of the nicotinamide SAM methyltransferase inhibitor, developing an inhibitor which binds to the substrate binding site as well as to the cofactor binding site could be effective. As Nsp14 and Nsp16 target different substrates, any inhibitors designed in this way would likely have specificity to only one of the two methyltransferases from SARS-CoV-2. Of the two, Nsp14 might be easier to target as it has a unique structure not similar to human SAM methyltransferases.
As both Nsp14 and Nsp16 interact with Nsp10 for normal function, interfering with this interaction could reduce activity of these enzymes. Further still, as the interface between Nsp10 with Nsp14 and Nsp16 has overlap the target is smaller for blocking binding of these proteins.
One way to look for possible drugs is repurposing those which are already approved for other diseases. Initial screen can be done in silico, by simulations of the interaction between the protein and the already existing and approved drug. However, such studies are highly dependent on the protein structures employed being correct, which is why we are evaluating all structures that are published for SARS-CoV and SARS-CoV-2.
Available structures
If you would like to look at the currently available structures for Nsp10, Nsp14, and Nsp16, they are available from our data base; we provide information on the quality of measurement data and models as well as improved structures.
All structures available for Nsp14 are bound to Nsp10 and are only available from SARS-CoV. The highest resolution structure of Nsp14 is PDB entry 5c8t at 3.2Å. It has a bound S-Adenosyl methionine ligand as well as zinc ions present. Alongside this, another structure of Nsp14 bound to S-Adenosyl homocysteine and a guanosine-triphosphate-adenosine ligand as well as zinc at 3.33Å resolution has been published (PDB: 5c8s). Additionally, two structures with zinc atoms but no ligands are available (PDB 5c8u 3.4Å at and 5nfy at 3.34Å). Both PDB entries 5c8t and 5nfy have been improved structures by our group.
Similar to Nsp14 all structures of Nsp16 are bound to Nsp10. There are currently 18 structures for Nsp16 bound to Nsp10 from SARS-CoV-2. The highest resolution structure is at 1.8Å and has SAM, Guanosine triphosphate and Adenosine bound as well as zinc atoms. The PDB:6wkq has Nsp16 bound to the methyltransferase inhibitor Sinefungin at 1.98Å resolution. Two further structures of note are 7jhe and 7jib that have various functional ligands. A further four structures are available from SARS-CoV.
Nsp10 alone: Currently there are two structures of Nsp10 from SARS-CoV-2, PDB 6zpe and 6zct, with the former having the highest resolution of 1.58 Å with bound zinc (PDB 6zpe). There are also three structures of Nsp10 from SARS-CoV available, PDB 2fyg, 2g9t, and 2ga6.
- 1.Ramanathan A, Robb GB, Chan S-H. mRNA capping: biological functions and applications. Nucleic Acids Res. Published online June 17, 2016:7511-7526. doi:10.1093/nar/gkw551
- 2.Boriack-Sjodin PA, Swinger KK. Protein Methyltransferases: A Distinct, Diverse, and Dynamic Family of Enzymes. Biochemistry. Published online December 22, 2015:1557-1569. doi:10.1021/acs.biochem.5b01129
- 3.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. Published online October 16, 2017:81-92. doi:10.1038/nrg.2017.80
- 4.Krafcikova P, Silhan J, Nencka R, Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun. Published online July 24, 2020. doi:10.1038/s41467-020-17495-9
- 5.Ferron F, Subissi L, Silveira De Morais AT, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci USA. Published online December 26, 2017:E162-E171. doi:10.1073/pnas.1718806115
- 6.Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc Natl Acad Sci USA. Published online July 9, 2015:9436-9441. doi:10.1073/pnas.1508686112
- 7.Rosas-Lemus M, Minasov G, Shuvalova L, et al. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. Published online April 20, 2020. doi:10.1101/2020.04.17.047498
- 8.Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells. Published online May 20, 2020:1267. doi:10.3390/cells9051267
- 9.Chen Y, Su C, Ke M, et al. Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex. Kuhn RJ, ed. PLoS Pathog. Published online October 13, 2011:e1002294. doi:10.1371/journal.ppat.1002294
- 10.Robert L. H, Marvin M. H. A9145, A NEW ADENINE-CONTAINING ANTIFUNGAL ANTIBIOTIC. J Antibiot. 1973;26(8):463-465. doi:10.7164/antibiotics.26.463
- 11.Policarpo RL, Decultot L, May E, et al. High-Affinity Alkynyl Bisubstrate Inhibitors of Nicotinamide N-Methyltransferase (NNMT). J Med Chem. Published online October 7, 2019:9837-9873. doi:10.1021/acs.jmedchem.9b01238
The genome of the novel SARS-CoV-2 codes for an ORF1a/ ORF1ab (open reading frame) polyprotein containing sixteen non-structural proteins (NSP) and four structural proteins. The genome also has multiple ORFs coding for accessory proteins through a frame shift. These accessory proteins are not necessary for viral replication but might play a key role in pathogenesis of SARS-CoV-2. One such protein is the accessory protein 7a, which is predicted to contribute to Covid-19 by inducing the apoptotic processes in human host cells1.
Structure
SARS-CoV-2 is a very young virus and the structure and function of the accessory protein 7a has not yet been solved. However, 7a of SARS-CoV-2 shows 85% sequence identity and 95.2% sequence similarity with another protein in SARS-CoV2. It is therefore conceivable that both accessory proteins have a similar structure and function. The sequence analysis of SARS-CoV predicts that ORF7a codes for a type I transmembrane protein with 122 amino acids, including a signal peptide at the N‑terminus and a retrieval signal at the C-terminus3. The N-terminal ectodomain of ORF7a consists of seven β-strands compactly arranged in an immuno-globulin-like β-sandwich fold (Fig 1). These seven β-strands are ordered in two β-sheets containing four β-strands (A; G; F; C) in the first sheet and three (B; E; D) in the second one (see Fig 1: left)4.
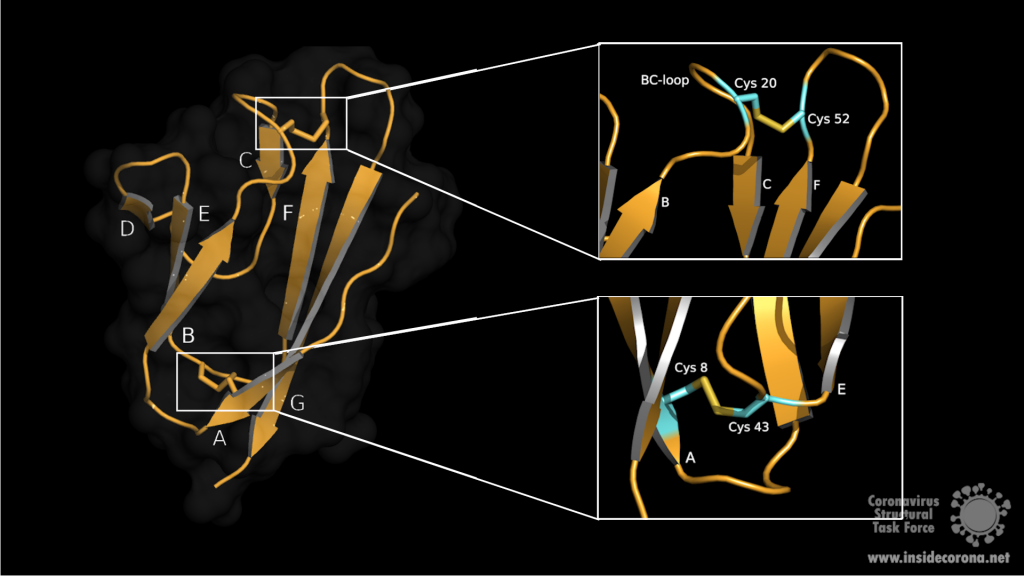
Both sheets are amphipathic, with the hydrophobic side facing inwards closely packed against each other. The top of the ectodomain is defined by the BC, DE and FG loops and the bottom by the AB, CD and EF loops. The β-sandwich structure is stabilized by two disulphide bonds linking the sheets at opposite edges. At the bottom of the structure, a disulphide bridge connects Cys8 on strand A with Cys43 at the end of strand E. At the top, Cys20 of the BC loop is linked to Cys52 at the end of strand F (see Fig 1: right). Additionaly, on top of the BED sheet, the DE loop protrudes from the structure and forms a groove together with β-strands C and D. In the centre is Glu18 which contributes to the negatively charged bottom of the mainly hydrophobic groove. This grove may be a potential site for ligand interaction due to its central negative electrostatic potential4.
Function
In cell culture, the polypeptide 7a of SARS-CoV seems to have diverse biological functions5. It is possible that 7a plays a key role in cell cycle control. In HEK 293 cells, an overexpression of 7a led to inhibition of cell growth and induction of the G0/G1 phase cell cycle arrest. This arrest may favour coronavirus replication and exacerbate virus-induced pathogenicity. 7a is also predicted to induce apoptosis in human kidney epithelial cells by interaction with a protein called B-cell lymphoma-extra large (Bcl-XL). Bcl-XL belongs to a group of pro-survival proteins, the B-cell lymphoma-2 (Bcl-2)- family, which prevent apoptosis in epithelial cells. The Interaction between 7a and the C-terminal transmembrane domain of Bcl-XL may interfere with this pro-survival function, leading to apoptosis via the caspase-dependant pathway6,7. In addition to this, SARS 7a interacts with a Ap4A-hydrolase involved in cell proliferation, DNA-replication, apoptosis and RNA-processing. This interaction leads to downregulation of its hydrolase-activity and an increased production of AP4A (diadenosine tetraphosphate) which may also induce apoptosis5. Such a host cell specific modulation of apoptosis could enable the virus to evade the immune response or to spread to other target organs.
Another predicted function of ORF7a is the inhibition of the bone marrow matrix antigen 2 (BST-2) that might restrict virus release by physically tethering the budding enveloped virion to the plasma membrane. ORF7a antagonizes this function by binding of the extracellular domain of BST-2 preventing its glycosylation. Thus, an inhibitor preventing ORF7a-BST-2 interaction can be speculated as potential drug target8.
Taken together, ORF7a is a virulence factor that contributes in different ways to the pathogenicity of SARS-CoV-2. Therefore, targeted drug development against ORF7a could be a critical factor to reduce viral spread or attenuate severe disease progression.
PDB Structures Available
6W37: X-ray structure of the SARS-CoV-2 ORF7a encoded accessory protein.
1xak: SARS-CoV ORF7a accessory protein, a unique type I transmembrane protein of unknown function. Has a short cytoplasmic tail and a transmembrane domain. Consists of one chain (chain A), that forms a compact seven-stranded beta sandwich.
1y04: SARS Coronavirus ORF 7a coded X4 protein, also known as 7a, U122 or X4. Type-I transmembrane protein with immunoglobulin like beta-sandwich fold. Potential functions of X4 in virus replication and pathogenesis are discussed.
References
- 1.Michel CJ, Mayer C, Poch O, Thompson JD. Characterization of accessory genes in coronavirus genomes. Virol J. Published online August 27, 2020. doi:10.1186/s12985-020-01402-1
- 2.Francis K. Y. The Proteins of Severe Acute Respiratory Syndrome Coronavirus‑2 (SARS CoV‑2 or n‑COV19), the Cause of COVID‑19. The Protein Journal (2020). 2020;(39):198-216. doi:10.1007/s10930-020-09901-4
- 3.Fielding BC, Tan Y-J, Shuo S, et al. Characterization of a Unique Group-Specific Protein (U122) of the Severe Acute Respiratory Syndrome Coronavirus. JVI. Published online July 15, 2004:7311-7318. doi:10.1128/jvi.78.14.7311-7318.2004
- 4.Hänel K, Stangler T, Stoldt M, Willbold D. Solution structure of the X4 protein coded by the SARS related coronavirus reveals an immunoglobulin like fold and suggests a binding activity to integrin I domains. J Biomed Sci. Published online November 23, 2005:281-293. doi:10.1007/s11373-005-9043-9
- 5.Vasilenko N, Moshynskyy I, Zakhartchouk A. SARS coronavirus protein 7a interacts with human Ap4A-hydrolase. Virology Journal. Published online 2010:31. doi:10.1186/1743-422x-7-31
- 6.Tan Y-J, Fielding BC, Goh P-Y, et al. Overexpression of 7a, a Protein Specifically Encoded by the Severe Acute Respiratory Syndrome Coronavirus, Induces Apoptosis via a Caspase-Dependent Pathway. JVI. Published online December 15, 2004:14043-14047. doi:10.1128/jvi.78.24.14043-14047.2004
- 7.Tan Y-X, Tan THP, Lee MJ-R, et al. Induction of Apoptosis by the Severe Acute Respiratory Syndrome Coronavirus 7a Protein Is Dependent on Its Interaction with the Bcl-XL Protein. JVI. Published online April 11, 2007:6346-6355. doi:10.1128/jvi.00090-07
- 8.Taylor JK, Coleman CM, Postel S, et al. Severe Acute Respiratory Syndrome Coronavirus ORF7a Inhibits Bone Marrow Stromal Antigen 2 Virion Tethering through a Novel Mechanism of Glycosylation Interference. García-Sastre A, ed. J Virol. Published online September 16, 2015:11820-11833. doi:10.1128/jvi.02274-15
The building plan
Storing the building plans for a virus in its genome is much like how we store ideas in language. This may sound strange but, as an example, typos in spelling, grammar, or word usage, can lead to the meaning of a sentence either changing dramatically, remaining virtually unchanged, or becoming complete nonsense. The SARS-CoV-2 genome consists of RNA. Transcription of this RNA runs into a similar problem: errors can lead to the loss of function, a gain of function, or be completely inconsequential to the resulting protein (Figure 1). Large changes may break the virus, but smaller changes may provide an advantage and are essential for evolution.
Targeting the copy machine
In a previous article we spoke about the copy machinery of the virus, including the RNA-dependent RNA polymerase (RdRp), and drugs targeting it, such as Remdesivir. The goal of these drugs is to jam the enzyme and halt RNA production - or to cause more errors than are sustainable, with the end result being a less infectious virus. The reason the development of drugs targeting the copy machinery of RNA is worthwhile is that humans don’t have machinery to reproduce RNA from RNA. This means drugs targeting this machinery are less likely to interfere with normal processes in people. What if the virus could quickly repair these errors before the new genome is packed into a hull and kicked out the door? That would make finding a therapeutic much more difficult…
Correctional facilities
Unfortunately, SARS-CoV-2 has a way to repair the mistakes. When errors are introduced in transcription through environmental mutagenesis or even mutations caused by nucleotide analogs like Ribavarin1–3, the non-structural protein 14 (nsp14) has the ability to remove them. This multifunctional protein removes errors with the exoribonuclease (ExoN) activity of its N-terminal domain, while the C-terminal domain has the unrelated function of methylating the end cap of the viral RNA3,4.
However, this ExoN does not work alone. There is a replication complex made up of proteins performing many roles in the production of new RNA with high fidelity. Nsp12 is the main hub that makes a new RNA chain to complement the template. Nsp7 and nsp8 have a “processivity” role to enable nsp12 to function efficiently. In addition to these proteins there is a two-component proofreading system of Helicase (nsp13) and the ExoN domain of nsp14. Helicase can detect misshapen RNA helices caused by errors made by the copy machinery5. It then unwinds these double strands of RNA and feeds the strand containing the error into the ExoN domain of nsp14 where they are chopped out. This results in nsp12 continuing RNA replication where it left off.
Exoribonuclease or no exoribonuclease
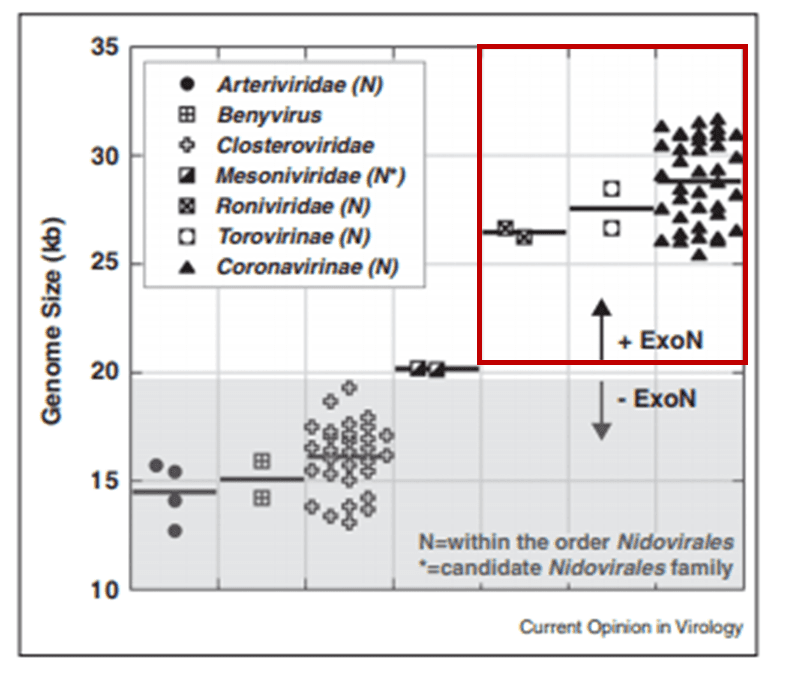
The proofreading ability from Helicase and nsp14 ExoN allows SARS-CoV-2 to have a huge genome as compared to other viruses6(Figure 2). The large 29.9 kb genome of SARS-CoV-2 requires much more physical space to accommodate the necessary genetic information for reproduction when compared to other RNA viruses, such as Rhinovirus that has a genome between 7.2 kb and 8.5 kb in size (Figure 3). When no ExoN proofreading is present genomes cannot expand beyond 20 kb in size6(Figure 2). Maybe by removing the exoribonuclease activity, irreversible damage could be caused to the genome of SARS-CoV-2.

Nsp14 Structure
In order to understand how nsp14 can do this, we need to find out its atomic structure; this may also allow us to develop a drug which hinders its function. However, to this date, no structure of nsp14 from SARS-CoV-2 has been solved. However, structures have been solved of nsp14 in complex with another viral protein, nsp10, both from SARS-CoV (PDB entries 5nfy, 5c8s, 5c8t, 5c8u)2,7. As the protein sequences are very similar between SARS-CoV and SARS-CoV-2 (nsp14 is 95%, and nsp10 is 97% identical), it can be assumed that the SARS-CoV-2 structure as well as its functionality are very similar to SARS-CoV. The active site of the ExoN domain of nsp14 from SARS-CoV-2 has a DEEDh motif (named for the one-letter codes of the amino acids involved) containing a histidine as well as two aspartates and two glutamates2,3,7,8.
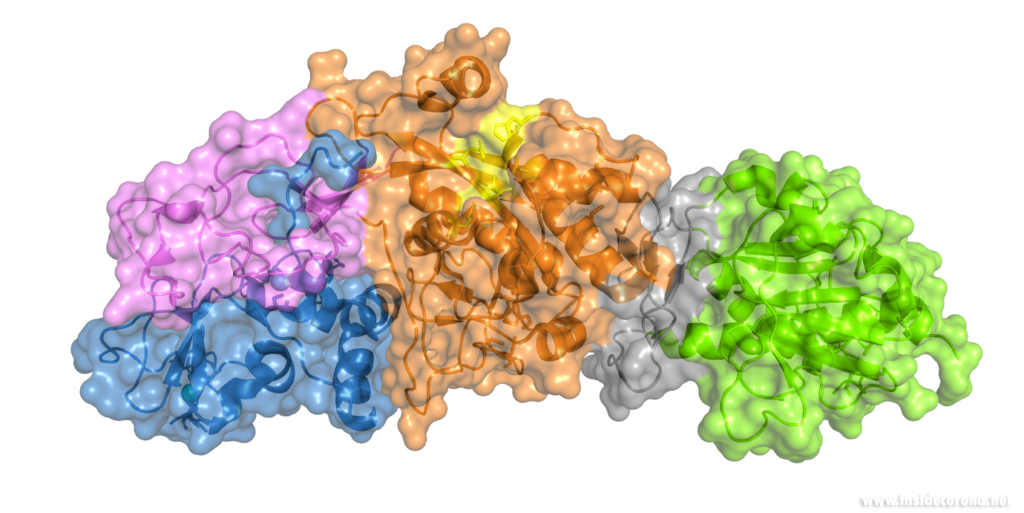
Nsp14 interacts with nsp10
The N-terminus of nsp14 interacts with nsp10 (pink and blue, respectively, in Figure 4). The following domain (orange) has been shown to have exoribonuclease activity on double stranded RNA in a 3’ to 5’ direction9. When nsp10 is interacting with nsp14 there is a 35 fold increase in exoribonuclease activity, which is thought to occur due to conformational changes caused by formation of the complex2,9. The ExoN domain of nsp14 (orange) is connected to the methyltransferase domain (green) by a flexible hinge (black)7,10. This flexible region opens up the methyltransferase active site to allow methylation of the N7 of the 5’ Guanosine triphosphate of RNA10. There are three zinc finger motifs in nsp14 with two found in the ExoN domain and one in the methyltransferase domain2,7. In combination with the two further zinc sites in nsp10, these zinc fingers hold loops of the proteins together and are involved with nucleotide interaction2,7.
Nsp14 has also been demonstrated to form complexes with the copy machinery , nsp12, nsp7, and nsp8, although this interaction is independent of nsp102,11,12.
Exoribonuclease active site and potential drug development
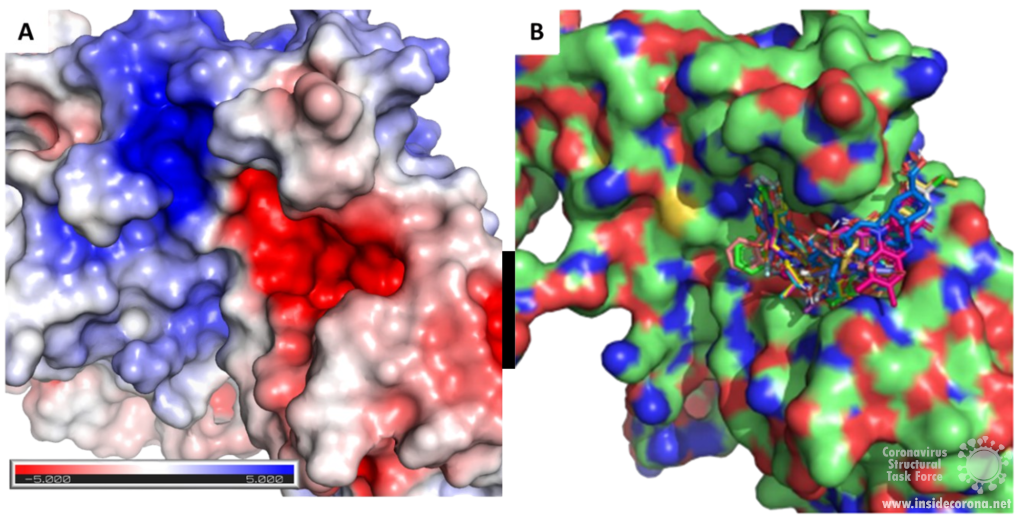
Scientists are searching for drugs that could be used to target nsp14 in order to find a cure for COVID-19. The active site of the ExoN domain of nsp14 has five residues that are essential for activity that form a negatively charged pocket (Figure 5A)7. Currently researchers are using the nsp14 structure from SARS-CoV to model a SARS-CoV-2 structure which can be used to identify compounds that could bind to the active site (Figure 5). These in silico screens start with nucleotide analog drugs like Remdesivir, Ribivarin or Ritonavir that are currently used as antiviral treatments for other viruses13–15. These nucleotide analogs are then changed to achieve a better binding to Nsp14’s active site in order to block it (Figure 5B).
As the ExoN is essential to support the huge 29.9kb genome of SARS-CoV-2, targeting nsp14 could lead to an effective treatment to COVID-19. Although drugs that target just nsp14 could be effective at increasing the error rate in RNA production by the virus, a more effective treatment will require inhibition of the RdRp of the copy machinery at the same time!
Available structures
If you would like to look at the currently available structures for Nsp14(currently only available from SARS-CoV), they are available from our data base; we provide information on the quality of measurement data and models as well as improved structures. The highest resolution structure of nsp14 is PDB entry 5c8t at 3.2Å. This has a bound S-Adenosyl methionine ligand as well as zinc atoms present. Alongside this, another structure of Nsp14 bound to S-Adenosyl homocysteine and a guanosine-triphosphate-adenosine ligand as well as zinc at 3.33Å resolution has been published (PDB: 5c8s). Additionally, two structures with zinc atoms but no ligands are available (PDB 5c8u 3.4Å at and 5nfy at 3.34Å). Both PDB entry 5c8t and 5nfy have improved structures re-refined by our group.
Sources
- 1.Zuo Y. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Research. Published online March 1, 2001:1017-1026. doi:10.1093/nar/29.5.1017
- 2.Ferron F, Subissi L, Silveira De Morais AT, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci USA. Published online December 26, 2017:E162-E171. doi:10.1073/pnas.1718806115
- 3.Barnes MH, Spacciapoli P, Li DH, Brown NC. The 3′–5′ exonuclease site of DNA polymerase III from Gram-positive bacteria: definition of a novel motif structure. Gene. Published online January 1995:45-50. doi:10.1016/0378-1119(95)00530-j
- 4.Chen Y, Cai H, Pan J, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proceedings of the National Academy of Sciences. Published online February 10, 2009:3484-3489. doi:10.1073/pnas.0808790106
- 5.Chen J, Malone B, Llewellyn E, et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Published online July 8, 2020. doi:10.1101/2020.07.08.194084
- 6.Smith EC, Denison MR. Implications of altered replication fidelity on the evolution and pathogenesis of coronaviruses. Current Opinion in Virology. Published online October 2012:519-524. doi:10.1016/j.coviro.2012.07.005
- 7.Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc Natl Acad Sci USA. Published online July 9, 2015:9436-9441. doi:10.1073/pnas.1508686112
- 8.Eckerle LD, Becker MM, Halpin RA, et al. Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing. Emerman M, ed. PLoS Pathog. Published online May 6, 2010:e1000896. doi:10.1371/journal.ppat.1000896
- 9.Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proceedings of the National Academy of Sciences. Published online May 25, 2012:9372-9377. doi:10.1073/pnas.1201130109
- 10.Ogando NS, Ferron F, Decroly E, Canard B, Posthuma CC, Snijder EJ. The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity. Front Microbiol. Published online August 7, 2019. doi:10.3389/fmicb.2019.01813
- 11.Subissi L, Posthuma CC, Collet A, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA. Published online September 2, 2014:E3900-E3909. doi:10.1073/pnas.1323705111
- 12.Subissi L, Imbert I, Ferron F, et al. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: Replicative enzymes as antiviral targets. Antiviral Research. Published online January 2014:122-130. doi:10.1016/j.antiviral.2013.11.006
- 13.Khater S, Dasgupta N, Das G. Combining SARS-CoV-2 proofreading exonuclease and RNA-dependent RNA polymerase inhibitors as a strategy to combat COVID-19: a high-throughput in silico screen. Published online June 24, 2020. doi:10.31219/osf.io/7x5ek
- 14.Shannon A, Le NT-T, Selisko B, et al. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Research. Published online June 2020:104793. doi:10.1016/j.antiviral.2020.104793
- 15.Narayanan N, Nair DT. Ritonavir May Inhibit Exoribonuclease Activity of Nsp14 from the SARS-CoV-2 Virus and Potentiate the Activity of Chain Terminating Drugs. chemrxiv.org. Published May 13, 2020. https://chemrxiv.org/articles/Ritonavir_May_Inhibit_Exoribonuclease_Activity_of_Nsp14_from_the_SARS-CoV-2_Virus_and_Potentiate_the_Activity_of_Chain_Terminating_Drugs/12280043
Introduction
Have you heard that the coronavirus “mutates”? Or that there are “several strains” of it around the world? Sounds scary, right? However, the reality is that everything “mutates”. All organisms, over time, acquire differences in their genes, from bacteria to humans. You might be aware that this can happen when your DNA (Deoxyribonucleic Acid) is exposed to UV light (like from the sun!), but this can also happen during DNA replication. This is when a cell uses the template of one of the two DNA strands to make a new complimentary copy of the other strand. Mutation is common to all living organisms (and viruses) and a driver of evolution. This is the first post in a series that will explore coronavirus replication with a focus on the proteins involved.
How does the coronavirus make more of itself?
SARS-CoV-2 uses single-strand Ribonucleic acid (RNA) to encode its genome, not DNA, and hence belongs to a class of “single-strand RNA viruses”. For this reason, the virus needs a different way to copy its genome than “normal” cells have. The viral protein that copies the RNA is called an “RNA-dependent RNA polymerase” (RdRp). This protein uses the viral RNA as a template to make a new copy of viral RNA, by stringing single ribonucleotides together like beads on a string. This process is called polymerization.
A study by the Morse lab at Texas A&M University showed that SARS-CoV-2 RNA polymerase has a remarkable similarity to the RNA polymerase of SARS-CoV (>95%) as well as MERS-CoV [1], the virus which causes Middle-Eastern Respiratory Syndrome. This means that research performed in response to the SARS and MERS epidemics can inform our response to SARS-CoV-2. Unfortunately, a lack of consistent pandemic-preparedness funding means that we didn’t learn as much about RdRp in time as we could have. Still, RNA polymerase might be a viable drug target for halting the spread and reducing the fatality rate of COVID-19.
Structure of the RNA-Dependent RNA Polymerase
By determining the structure of RdRp, and deeply understanding how it works, we can optimize a drug to specifically target it and hinder its function. To this end, in the last few months, several structures of SARS-CoV-2 RNA polymerase have been published.
One interesting structure shows RNA polymerase in action, in the process of elongating an RNA strand (see Figure 1).[2] This structure clearly show the polymerase in complex with smaller proteins, non-structural protein 7 and 8 (nsp7 and nsp8). These proteins improve how well the RNA polymerase binds the template RNA and also how long it stays bound before dissociating – a feature called “processivity”.[3]
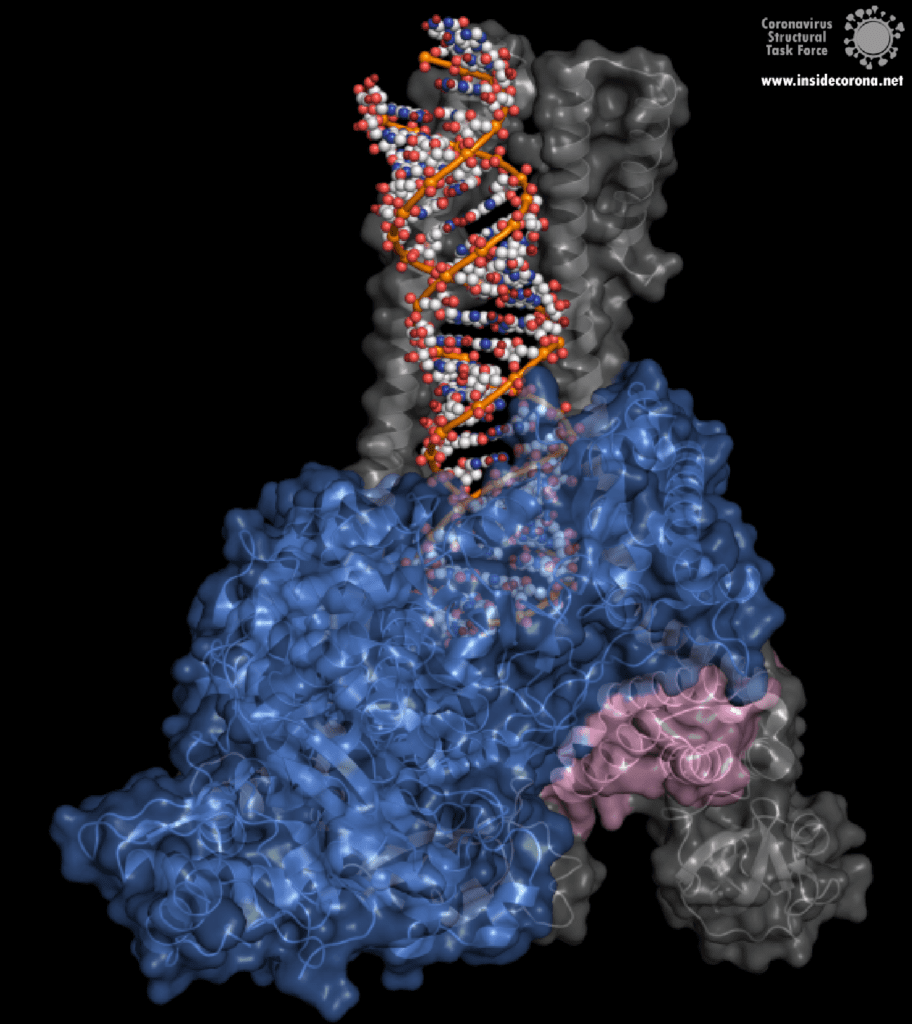
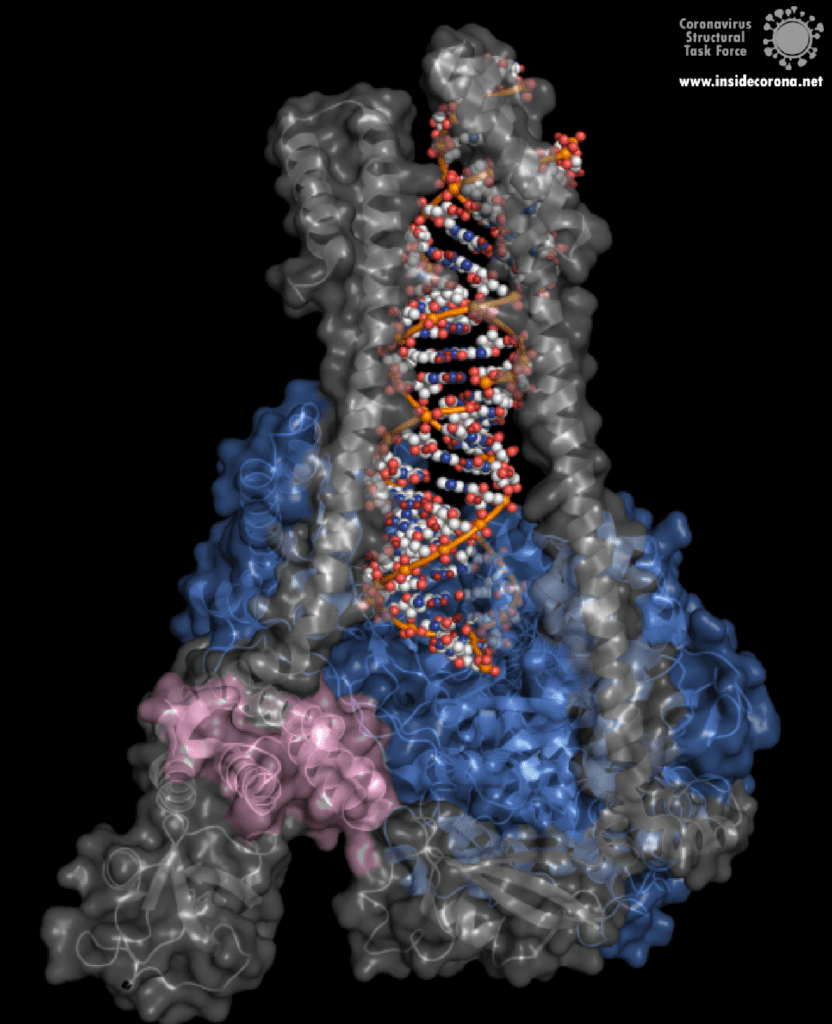
In the center of the protein is the area where the main action happens, called the “active site”. The amino acids of the polymerase that form the active site have a particular shape and chemical properties, which enable the polymerization reaction to occur very rapidly. In fact, the polymerase can string together as many as 100 nucleotides per second! [3] New RNA molecules can enter the active site through a little window to be added to the growing RNA chain. It is here that the antiviral drugs make their move!
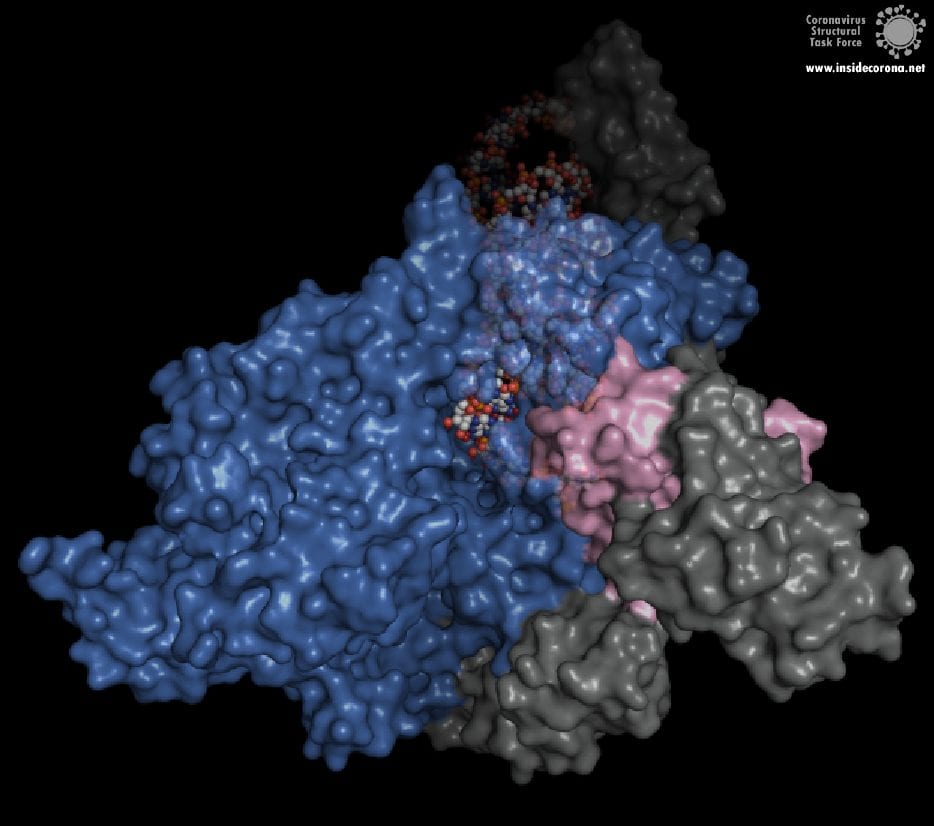
How do antiviral drugs attack RNA-dependent RNA polymerase?
First, let’s talk about Gilead’s FDA-approved drug, Remdesivir, which has taken the spotlight in the search for COVID-19 cures. Remdesivir (which has a fancy chemistry ID, GS-5734, and is sold under the brand name Veklury), is a “nucleotide analog”, which means that it mimics the shape and chemistry of the nucleotides that make up RNA and DNA (see figure).
Remdesivir was developed originally as a general antiviral drug and was later shown to protect cells (in a test tube) and monkeys (not in a test tube) from the Ebola Virus [4]. However, this was recent enough, and science is slow enough that, until the COVID-19 pandemic, large-scale clinical trials of Remdesivir hadn’t been done yet. So scientists and doctors have been rushing to test the drug in COVID-19 patients. In fact, the US and Japan both approved the drug for “Emergency Use Authorization'' for severe COVID-19 patients as early as May [5], [6]. And, in July, the European Medicines Agency gave Remdesivir a “conditional marketing authorization” (used for drugs that meet an unmet medical need but have insufficient data for normal approval). This allows the use of Remdesivir in severe COVID-19 patients through the next year [7]. So, how the heck does a drug for Ebola, Influenza, or some other viruses also work against COVID-19? I was concerned by this when the news about all the drug trials were coming out – and I’m sure I wasn’t the only one...
The simple answer to that is all these viruses need to do the same thing - copy their RNA genome from an RNA template. And in order to do that, they all end up using basically the same tool, an RNA-Dependent RNA polymerase. And all drugs that are nucleotide analogs use the very same trick: they dress up like ribonucleotides (the "beads on a string" from before) and fool the RNA polymerase into letting them into the active site. Once inside, they get “stuck” in the active site, jamming the polymerase machine. Since this trick should work for any viral RNA polymerase, we can use these drugs for any RNA virus, and call them ‘general antivirals’. Of course, in practice, this doesn't always work, because there are differences between the different RNA polymerases. However, it is a great place to start! In the future, if we have general antivirals for SARS-CoV-2 all ready-to-go, we may be better equipped to deal with another coronavirus outbreak!

The Chemistry of Remdesivir
Remdesivir resembles the nucleotide adenine in structure, although it has some fancy chemical add-ons which help make it a better drug (thank you, medicinal chemistry!). When Remdesivir is injected into a vein, it travels through the bloodstream and enters into our cells, which recognize it as a foreign substance and try to digest it. However, what ends up happening is that the cells remove just the fancy chemical add-ons, and then confuse it for a normal adenine nucleotide. In infected cells, the viral RNA-dependent RNA polymerase then starts grabbing these molecules and inserting them into the new viral RNA strand in place of adenine molecules. Remdesivir, now attached to the RNA, jams the polymerase, rendering the virus unable to make more copies of its genome. Ultimately, this halts viral replication and helps the patient fight off the virus.
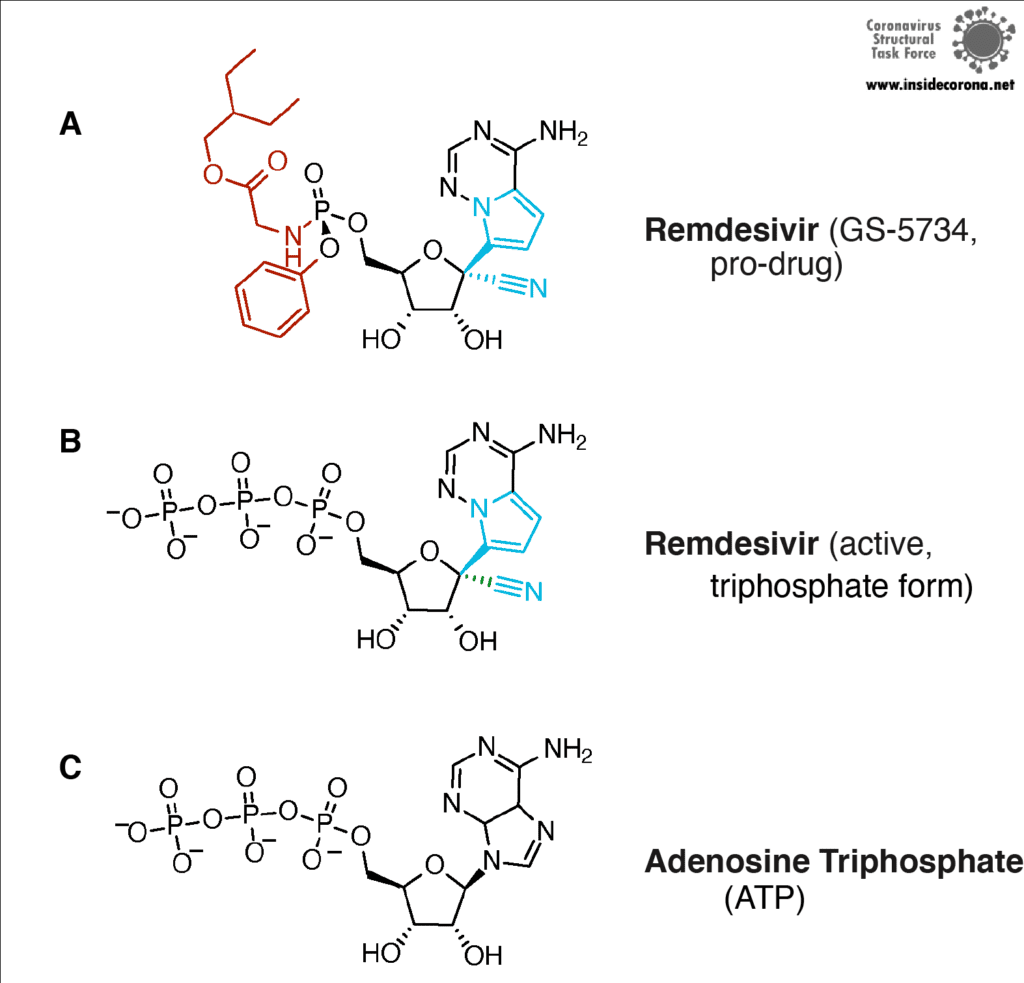
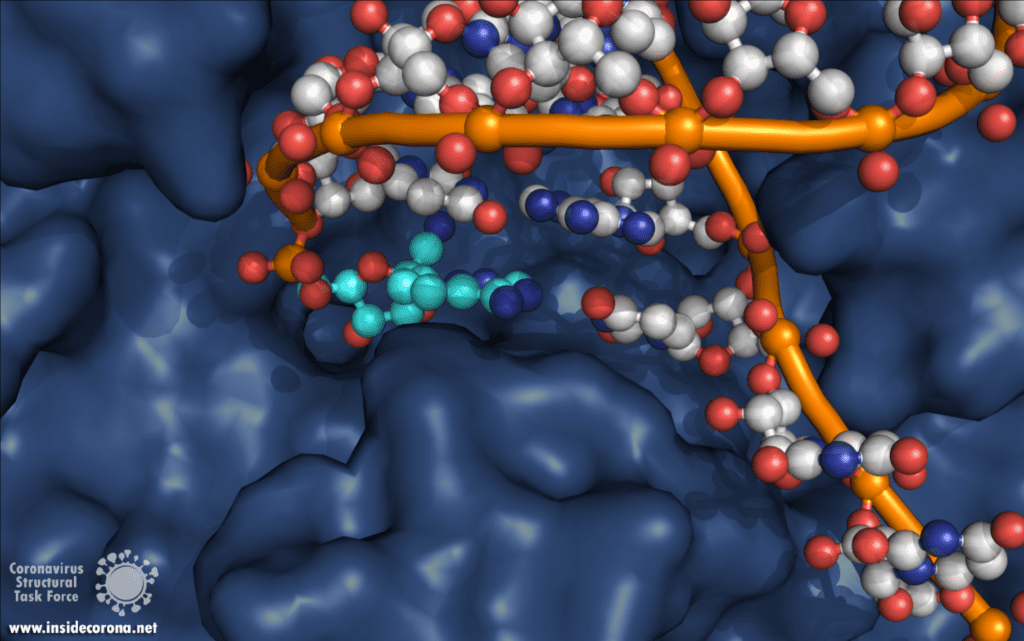
Another drug that inhibits the RNA polymerase activity is Favipiravir, sold under brand names Avigan, Abigan, and FabiFlu. Favipiravir has been discovered by Toyama Chemical Co., Ltd. in Japan and it has a similar mechanism to Remdesivir, except that it mimics a guanosine nucleoside instead of an adenine nucleotide [8]. This drug was approved in Japan back in 2014 for use in resistant cases of Influenza A and B, but still remains unapproved in the US (still in Phase II and Phase III clinical trials) and the UK [9]. This drug is also being tested for use against Ebola virus, Lassa virus, and currently SARS-CoV-2 in 43 countries. The approval of Favipiravir for COVID-19 has been much faster in China (Mar 15, 2020), Russia (Jun 3, 2020), and India (Jun 20, 2020)[10], [11]. Nonetheless, other countries, including Japan, are in various stages of clinical trials, and the results are anticipated to be out by the end of July [10].
So...do we have a cure for SARS-CoV-2?
Sadly, not yet. While the speed at which Remdesivir has gone through clinical trials is unprecedented, more work needs to be done to make sure it is safe and effective. Since (in the big scope of things) not a lot have people have taken Remdesivir, we aren’t really sure what all the side effects are, although there is emerging evidence for liver and kidney damage [12, 13]. The most common side effects are nausea (10% and 9% of patients), indigestion (7%) and increase of transaminases (6% and 8%). In one study, 3.6% of patients in a 10-day trial needed to stop taking therapy due to the latter. However, serious viral infections can also cause liver damage, so separating the two causes is a challenge! Remdesivir is not a cure-all, either. In one study it improved the recovery time from 15 days to 11 days, but it showed no effect for patients with mild to moderate disease, and no difference in median recovery time for patients who were already on a ventilator [14]. Since the drug has to be given by infusion over several days, there is a pretty small window in which Remdesivir can actually help.
Likewise, Favipiravir has its own side effects such as liver damage, elevated uric acid levels, kidney damage, skin allergies, etc. [15]. These effects restrict it for use by severe diabetes and heart patients. On top of that, it is not suitable for pregnant women because it can cause potential fetal deaths and deformities. It has been shown that Favipiravir works only during the earlier stages of SARS-CoV-2 infection when the body’s immune system isn’t totally drained, whereas it can result in a cytokine storm (when your immune system really freaks out) in severely ill patients. But, unfortunately, the virus doesn’t differentiate between humans while attacking, so a universal drug for COVID-19 has to be safe for use by all people.
However, these drugs are better than nothing, and by understanding the mechanisms involved, scientists can continue to improve upon the existing drugs for the benefit of all. While most of the ‘general antivirals’ that target RNA Polymerase have failed with SARS-CoV-2, Remdesivir has been relatively successful. Scientists think that this is actually because of a proofreading protein in SARS-CoV-2 called exonuclease. Immediately after the RNA-polymerase makes new RNA, exnuclease checks to make sure the new RNA is correct. In one study, another drug that mimics RNA called Ribivarin was shown to be removed from newly synthesized RNA by exonuclease [16]. Thankfully, Remdesivir is not excised , which is likely why it has been more successful than the other options [17], [18]. To read more about how nsp14 maintains the integrity and virulence of SARS-CoV-2, tune in to a future blog entry!
Recommended Structures
For those interested in reviewing the structures further, they are available in our GitHub repo, along with information about validation and, where relevant, improved structures. For a high-resolution comparison of the active site with and without Remdesivir, 7BV2 and 7BV1 (respectively) were published together at 2.5 and 2.8 Å. The elongating structure of the complex shown above (6YYT) has the polymerase as well as the cofactors and RNA very well resolved, with little "missing" density and a resolution of 2.9 Å. It is likely preferable to 6M71 and 7BTF, which were published with a similar resolution but with less of the complex resolved, and no RNA. For those interested, 7C2K and 7BZF (at 2.93 Å and 3.26 Å) show the complex bound to RNA in a pre- and post-translocation state.
Sources
[1] J. S. Morse, T. Lalonde, S. Xu, and W. R. Liu, “Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV,” ChemBioChem, vol. 21, no. 5, pp. 730–738, Mar. 2020, doi: 10.1002/cbic.202000047.
[2] H. S. Hillen, G. Kokic, L. Farnung, C. Dienemann, D. Tegunov, and P. Cramer, “Structure of replicating SARS-CoV-2 polymerase,” Nature, May 2020, doi: 10.1038/s41586-020-2368-8.
[3] W. Yin et al., “Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir,” Science, p. eabc1560, May 2020, doi: 10.1126/science.abc1560.
[4] R. T. Eastman et al., “Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19,” ACS Cent. Sci., May 2020, doi: 10.1021/acscentsci.0c00489.
[5] O. of the Commissioner, “Coronavirus (COVID-19) Update: FDA Issues Emergency Use Authorization for Potential COVID-19 Treatment,” FDA, May 04, 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment (accessed Jul. 08, 2020).
[6] A. Sternlicht, “Japan Approves Remdesivir For Use On Severe COVID-19 Patients,” Forbes. https://www.forbes.com/sites/alexandrasternlicht/2020/05/07/japan-approves-remdesivir-for-use-on-severe-covid-19-patients/ (accessed Jul. 08, 2020).
[7] D. CZARSKA-THORLEY, “First COVID-19 treatment recommended for EU authorisation,” European Medicines Agency, Jun. 25, 2020. https://www.ema.europa.eu/en/news/first-covid-19-treatment-recommended-eu-authorisation (accessed Jul. 10, 2020).
[8] E. De Clercq, “New Nucleoside Analogues for the Treatment of Hemorrhagic Fever Virus Infections,” Chem. Asian J., vol. 14, no. 22, pp. 3962–3968, Nov. 2019, doi: 10.1002/asia.201900841.
[9] K. Shiraki and T. Daikoku, “Favipiravir, an anti-influenza drug against life-threatening RNA virus infections,” Pharmacol. Ther., vol. 209, p. 107512, May 2020, doi: 10.1016/j.pharmthera.2020.107512.
[10] T. Hornyak, “Japan sending Fujifilm’s flu drug favipiravir to over 40 countries for Covid-19 trials,” CNBC, May 04, 2020. https://www.cnbc.com/2020/05/04/fujifilms-flu-drug-favipiravir-sent-to-43-nations-for-covid-19-trials.html (accessed Jul. 14, 2020).
[11] G. P. Ltd, “Glenmark Becomes the First Pharmaceutical Company in India to Receive Regulatory Approval for Oral Antiviral Favipiravir, for the Treatment of Mild to Moderate COVID-19.” https://www.prnewswire.com/in/news-releases/glenmark-becomes-the-first-pharmaceutical-company-in-india-to-receive-regulatory-approval-for-oral-antiviral-favipiravir-for-the-treatment-of-mild-to-moderate-covid-19-855346546.html (accessed Jul. 14, 2020).
[12] Goldman, J. D. et al. Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. N. Engl. J. Med. (2020) doi:10.1056/NEJMoa2015301
[13] Remdesivir Safety Forecast: Watch the Liver, Kidneys | MedPage Today. https://www.medpagetoday.com/infectiousdisease/covid19/86582
[14] J. H. Beigel et al., “Remdesivir for the Treatment of Covid-19 — Preliminary Report,” N. Engl. J. Med., vol. 0, no. 0, p. null, May 2020, doi: 10.1056/NEJMoa2007764.
[15] Sandhya Ramesh, “Favipiravir, Japanese drug that’s the new Covid treatment hope your chemist will soon stock,” ThePrint, Jun. 25, 2020. https://theprint.in/health/favipiravir-japanese-drug-thats-the-new-covid-treatment-hope-your-chemist-will-soon-stock/447987/ (accessed Jul. 14, 2020).
[16] F. Ferron et al., “Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA,” Proc. Natl. Acad. Sci., vol. 115, no. 2, pp. E162–E171, Jan. 2018, doi: 10.1073/pnas.1718806115.
[17] C. J. Gordon, E. P. Tchesnokov, J. Y. Feng, D. P. Porter, and M. Gotte, “The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus,” J. Biol. Chem., Feb. 2020, doi: 10.1074/jbc.AC120.013056.
[18] L. Zhang et al., “Role of 1’-Ribose Cyano Substitution for Remdesivir to Effectively Inhibit both Nucleotide Addition and Proofreading in SARS-CoV-2 Viral RNA Replication,” bioRxiv, p. 2020.04.27.063859, Apr. 2020, doi: 10.1101/2020.04.27.063859.
The instructions and files below will allow you to create your own model of the virus! All you need is some spare time and a 3D printer. In addition, those without access to a 3D printer can still use the STL files to request printing from external services and then follow the instructions on painting and assembling the same way. We do hope that this model will make the virus more tangible, and that the model will not only be printed as a private project, but also be used for outreach activities and in educational institutions.
Our design is based on the best scientific evidence available. Not only are the shapes of the various proteins as true as we can make them, but their numbers as well as the overall size of the virion match experimental results on a scale of 1:1,000,000. If you want to know more about it, please look here. Once you have built a model from our design you will have a good representation of what one of these virions is expected to look like, after being scaled up by a factor of 1,000,000. Therefore 1 mm on the model represents 1 nm (10 Å). (By the way, this would make the RNA that is inside the virus hull 10 metres long and 1 mm thick, and the nucleocapsid around which the RNA is coiled would be about 1 metre and 1 cm in diameter.
We have also designed a scale model of the human anti-body that binds to the spike protein. This is available alongside the virus model and can be attached to the spike protein as desired. For easier printing, painting, and assembly, the virus structure has been broken down into 4 unique components:
To date the structures have been printed successfully on several Fused Deposition Modelling (FDM) printers (Rostok MAX v2 & Prusa I3 MK3 printers), and we anticipate the even higher quality structures will be feasible with alternate methods, such as stereolithography (watch this space). Let us know in the comments! Each of the parts is available in STL format and should be printable through any suitable slicer software. Personal discretion is advised when setting up the prints, as the exact details may differ depending on conditions and equipment. The procedure outlined below will serve as a good starting point.
Printing of the component parts
The first step is to print the individual components. For the virion parts this is very straight forward as the flat surface negates the need for supports. The virion objects can be printed with the minimum infill for support, though infill of 10% is recommended for rigidity.
The other parts (spike proteins and antibodies) provide a more challenging print. The spike protein must be printed 95 times to complete the model, and users can arrange these individually, or using 4 prints of 25x STL file. It is recommended that the spike protein is printed with the crown facing towards the print bed to maximize the support between the bed and eliminating the need to remove supports from the thin delicate stem.
A dual extruder printer would be ideal for spike printing as it would allow supports to be printed in a water soluble plastic, speeding up post-processing. In either case, printing individual or at least fewer spikes with greater spacing generally produces nicer objects that are easier to work with at the price of longer printing time. Indeed, there is a general trade-off between the convenience of the print set-up and the amount of post-processing and tidying needed for all 3D printing tasks, and one must find a compromise which satisfies them.
As stated above, we used FDM printing and ubiquitous poly-lactic acid (PLA), which made the post-processing easier.
Post-processing
Regardless of the approach taken for printing, some amount of tidying will typically be needed to get the objects ready for assembly. Removing the supports can be done with a pair of plyers, while the smaller artifacts and issues will need brushing off or sanding. A dental pick can be quite useful.
For PLA, we found the best thing to clean and smooth the surfaces (after support removal), is ethyl acetate. Ethyl acetate dissolves the plastic, breaking down the small extrusion artifacts on the surfaces. This can be used in many ways. We found it best to leave the parts in a sealed ethyl acetate vapour environment, such as a stainless steel pot, which should be cleaned carefully afterwards. This technique results in the most even and clean results, though will take up to a few days to fully smooth each object. The faster method, is to simply submerge the small objects in ethyl acetate for 10-30 seconds, and then remove each object, leaving them to dry out on a surface. For the larger virion parts, the surface can be smoothed by rubbing it down with a cloth damped with ethyl acetate. Ethyl acetate was also used to “weld” the two virion parts together. A small amount was dropped onto the flat surfaces on each section, before the two were pressed together until the plastic fused to become a single object. The seam was then smoothed down using the same process as before. Where one cannot get ethyl acetate from a lab or pharmacy, acetone-free nail-polish remover offers a commercially accessible alternative. you should be using safety glasses and suitable (!) gloves when handling ethyl acetate, ventilate the room well and if there was skin contact use a skin cream after hand washing.
It is worth noting that for the other common 3D printing material acrylonitrile butadiene styrene (ABS) or acetone may produce the same results.
Painting and Gluing

As with printing, painting methods and colours is down to personal preference, and here we outline our attempt, which was guided by the illustration by Thomas Splettstoesser as close as possible (see Fig. 3).
The parts were first treated with a primer to help the paint stick to the model. This also acts as a nice even Basecoat. When working with either primer or, as discussed later, an airbrush, one should consider safety: try to do as much as you can in a ventilated space, wearing safety goggles, gloves and a mask. Paint spraying produces a great number of fine particles which you don`t want to breathe in.
For us, the painting process was performed largely with an airbrush, and we highly recommend using one where available, due to the amount of painting required and surface complexity. Where not available, it can of course be done with just a simple brush which will take more time and a higher skill level.
All layer colours, medium thinner, base colours, primer and varnish we used were from Citadel painting. Here is an outline of the specific Citadel colours and materials we used for the model in the figures:
- Lime: “Moot green”
- Yellow: “Yriel Yellow”
- Grey: “Dawnstone”
- Wheat: “Baneblade Brown”
- Chocolate: “Doombull Brown”
- Aqua: “Gauss Blaster Green”
- Teal: “Kabalite Green”

The spikes were sorted into four sets in order to produce a graded lighting affect, with those on top brighter than those lower down. If you do not plan to use a base and do not have a fixed top and bottom you can skip this part.
We highlighted each Spike Protein with a brighter lime green to achieve more contrast to create depth, which makes the surface topology easier to distinguish. Finally, the highlighting of each spike was intensified by dry-brushing the protein with the “Aqua” colour.

After painting was complete the spikes and virion were sealed with gloss varnish and matte finish, respectively. This step is optional; however, the varnish protects the paints against damage and wear when being handled.
Finally, the 3D model was assembled. If highlighting was used in the painting step, one should ensure the spikes are placed so that brighter spikes go on top while darker ones at the bottom. Standard modeling glue was used to hold the spikes in place, though superglue or ethyl acetate would also work fine. Because we are planning on mounting this on a stand, we have left a hole at the bottom empty where the rod of our base will go in.
We hope that our adventure in 3D printing the Corona virus inspires you to give it a try! The process we described was completed in a little over a week. The printing jobs were completed in just over two days, the cleaning and post processing took another two days, while the painting was done over the course of a weekend. This article provides a description of our technique and should provide enough detail on how, with the outlined necessary tools, you could create a similar result. The files have been distributed through Thingiverse, and are distributed under a Creative Commons BY-NC license: You may remix, adapt, and build upon this work non-commercially and acknowledge the "Coronavirus Structural Task Force" as original author.

As with every 3D printed model, there are many different ways this could be tackled and achieved, and we look forward to seeing the many creative ways explored by others in this endeavor. Please do share experiences and results with us, either through the comments Thingiverse or on Twitter (you can tag us @thornlab or #insidecorona).
For a sense of perspective, we have also produced a model of the highly common rhinovirus, which is available in .stl format at the same scale as the corona virus objects. This is available at: https://www.thingiverse.com/thing:4556845.
Authors
We want to emphasize that the writing of this blog entry was a collaboration of a several people:
Dale Tronrud and Thomas Splettstoesser worked together to create the STL files for the 3D model. Dale was the person to suggest it first (with Andrea Thorn picking up on the idea). Thomas then selected the experimental models and placed all the parts to form a realistic representation. Dale provided the knowledge about the limitations imposed by the nature of 3D printing and broke up Thomas' model into printable parts that can be assembled without too much difficulty. He printed and assembled the first virion from this design.
Matt Reeves was responsible for improving the non-spherical virion model and the printing of the Würzburg model. He also determined the most suitable post-print processing techniques suitable for this project and, along with Dale and others on the team, contributed to many general technical discussions on how the model can be altered or improved further in the future.
Kristopher Nolte took part in the preprocessing and refining of the model together with Ferdinand Kirsten. Kristopher was also responsible for planning and carrying out the assembly and painting process of the Würzburg model.
Introduction
Before I started writing this article, the first thing I did was to google the name of my protein “NendoU” and was greeted by Figure 1. Needless to say, this is not what I was expecting. So, if you’re an anime fan looking for Riki Nendou, a dutiful yet dull-witted boy who likes helping people, particularly prioritising the weak, from The Disastrous Life of Saiki K: I’m afraid you have come to the wrong place. However, now that you’re here, maybe you’d like to learn about an interesting protein involved in SARS-CoV-2 viral replication? It can bind to and process six RNA molecules at a time! Six!
After that interlude, I should get this blog post back on track! So… viruses and proteins. SARS-CoV-2 is an enveloped coronavirus with a non-segmented positive-sense RNA genome, in English this means the RNA genome in SARS-COV-2 can be used “as is” to make viral proteins without prior modification. SARS-CoV-2 has one of the largest RNA genomes among RNA viruses, made up of a replicase gene encoding non-structural proteins (nsps), as well as various structural and accessory genes. During viral replication, depending on the starting point (a.k.a. a ribosomal frame shift), the replicase gene can produce one of two poly-protein chains, which are then cleaved to produce 15-16 individual viral nsps (non-structural Proteins). These nsps then form a large membrane-bound replicase complex with multiple enzymatic activities, like a tiny viral Voltron.
What’s in a Name?
This blog post will focus on SARS-CoV-2 Nsp15, a nidoviral RNA uridylate‐specific endoribonuclease (NendoU). That is a very long and complicated name which conveys a lot of information, so let’s break it down into its individual parts, like when Voltron separates to become several small robots. It’s possible I’ve watched too many cartoons during lockdown:
- Nidoviral – An order of RNA viruses which infect vertebrates and invertebrates.
- RNA – Genetic material used to produce proteins
- Uridylate-specific – Cuts Uridine (U) in RNA, not Cytosine (C), Adenine (A) or Guanine (G)
- Endo – A Greek word meaning inside or within
- Ribonuclease – An enzyme that cuts RNA into smaller pieces.
So, what’s in a name? Well, Nsp15 is a viral enzyme that likes to cut at uridine (a building block of RNA) in the middle of an RNA sequence. Quite a lot really. The final bit of the name “NendoU” goes into even more specifics on our protein, as it defines a common family of proteins which share certain traits. The first is that when Nsp15 cuts RNA, it gives a 2′‐3′ cyclic phosphodiester and 5′‐hydroxyl terminus. If we look at Figure 2, you’ll see a purple RNA chain made of two bases linked by an orange phosphate in the middle. When RNA is cleaved by Nsp15, a 2′‐3′ cyclic phosphodiester is made: in the two resulting molecules, the phosphate ion has been incorporated into a 5-membered ring (orange), and the other half of the RNA has a 5′‐hydroxyl, or and OH- group on another 5-membered ring (green). The second thing being a member of the NendoU family tells us is that the catalytic domain of the protein (the business end) is found on the C-terminal end of the protein (the latter half) as this is a shared trait within the NendoU family.
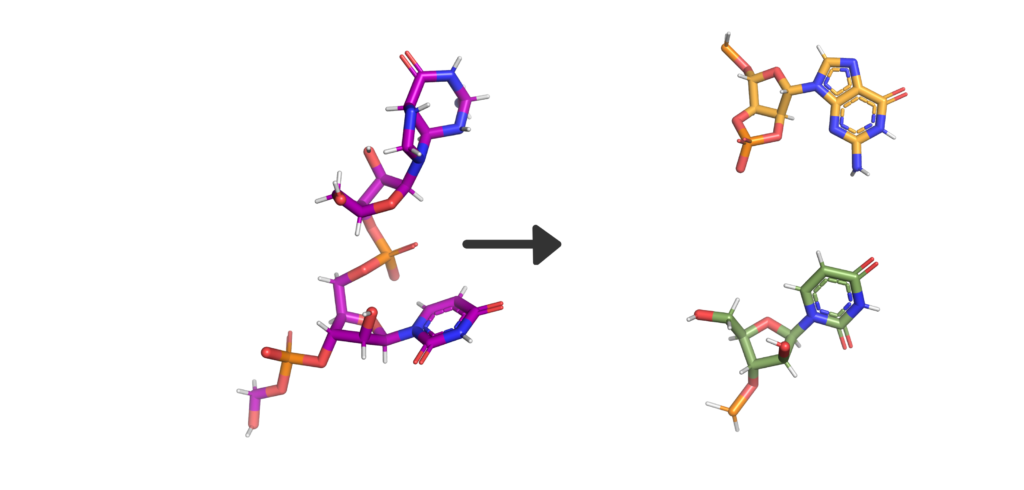
Domains
One Nsp15 monomer is made up of three distinct domains, the aforementioned N-terminal oligomerisation domain (green), a middle domain in… well, the middle (orange), and the catalytic NendoU domain at the C-terminal (purple, Figure 3b). Overall SARS-CoV-2 Nsp15 shows high sequence identity with SARS-CoV Nsp15 (88%) and, somewhat lower identity with MERS-CoV (51%) (Youngchang 2020), but the overall structural similarity is very high between the three viruses. For a more detailed breakdown of the secondary structure that makes up individual Nsp15 domains, check out our proteopedia entry!
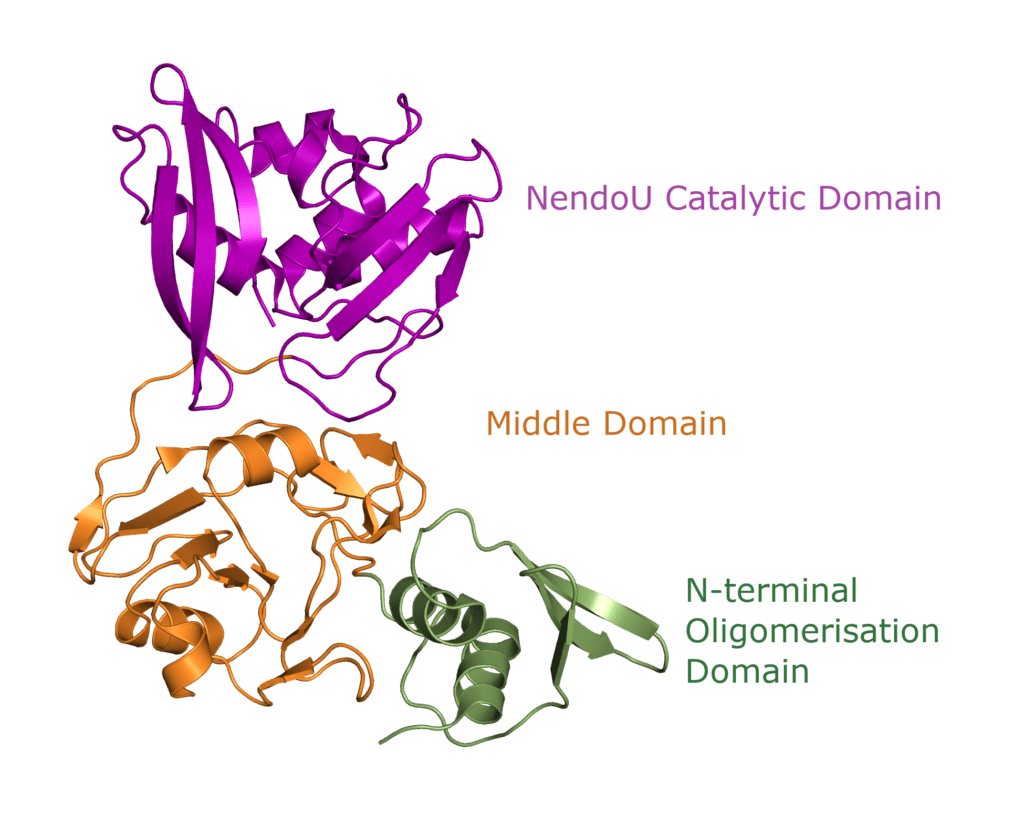
Tertiary Structure
Nsp15 forms a double-ring hexamer made up of a dimer of trimers stabilised by an N-terminal oligomerisation domain. So, three monomers form a trimer which then binds another trimer of monomers. However, If you open a crystal structures this can be confusing as you might not be presented with the whole complex. A crystal is composed of an infinite array of identical (or near enough) molecules related to each other by symmetry. To eliminate the need to store an infinite number of atoms on your computer the PDB file gives you just enough of the crystal to define the unique part. You are then expected to remember that the rest are generated by symmetry. This subset is called the asymmetric unit. Should you want to try and generate the whole crystal you can try, but your computer will likely grind to a halt on its way to infinity (and beyond).
For most structures the asymmetric unit is the interesting part. Often, when the biologically relevant complex has symmetry itself, like Nsp15 does, only part of the complex will be present in the file from the PDB. In the case of the PDB model 6X4I the molecules of each trimer obey the crystal’s three-fold symmetry. The file you download contains two molecules, one monomer from each trimer, and you must generate the symmetry related molecules (shown in green and orange in figure 3) to build the entire complex. These six monomers all come together to form the active enzyme, a 100 Å long and 10-15 Å wide channel, open to solvent from the top, bottom, and three separate side openings in the middle of the hexamer (Figure 4). Formation of the hexamer has been shown to be essential for enzymatic activity, making the oligomerisation interfaces a potential target for structure-based drug design. I’m not sure if I should be proud or disappointed that I didn’t mention Voltron once back there.
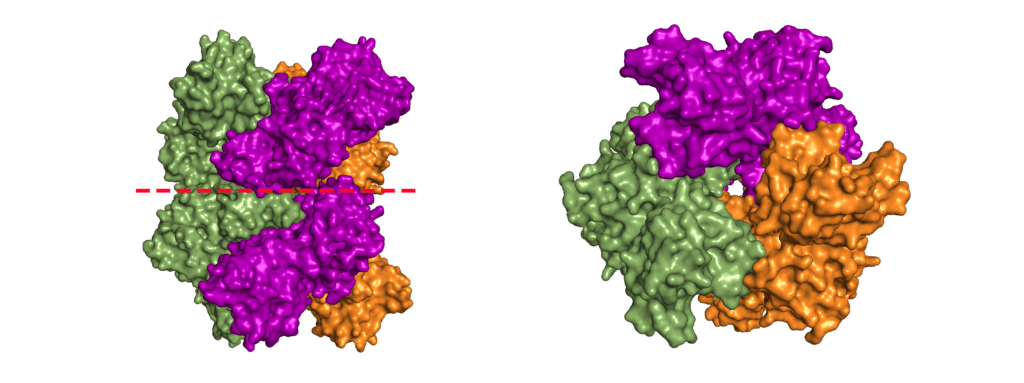
The Active Site
SARS-CoV-2 Nsp15 is a Mn2+ dependent endoribonuclease, meaning it relies on the coordination of manganese to perform the transesterification reaction (cutting RNA). Unfortunately, the structure of SARS-CoV-2 Nsp15 has not been solved with manganese present, but we do have a structure with 3’ uridine monophosphate in the active site (PDBID: 6X4I). It has been proposed that the presence of manganese help stabilise the active site and substrate, but it is yet to been seen. Based on sequence alignment against related enzymes from other viruses we know the active site is made up of six conserved residues that sit in a shallow groove between two β-sheets (His235, His250, Lys290, Thr341, Tyr343, and Ser294), as shown in Figure 5. His235, His250, and Lys290 are predicted to act as a catalytic triad, His235 as a general acid, and His250 as a base with Lys290 governing U specificity.
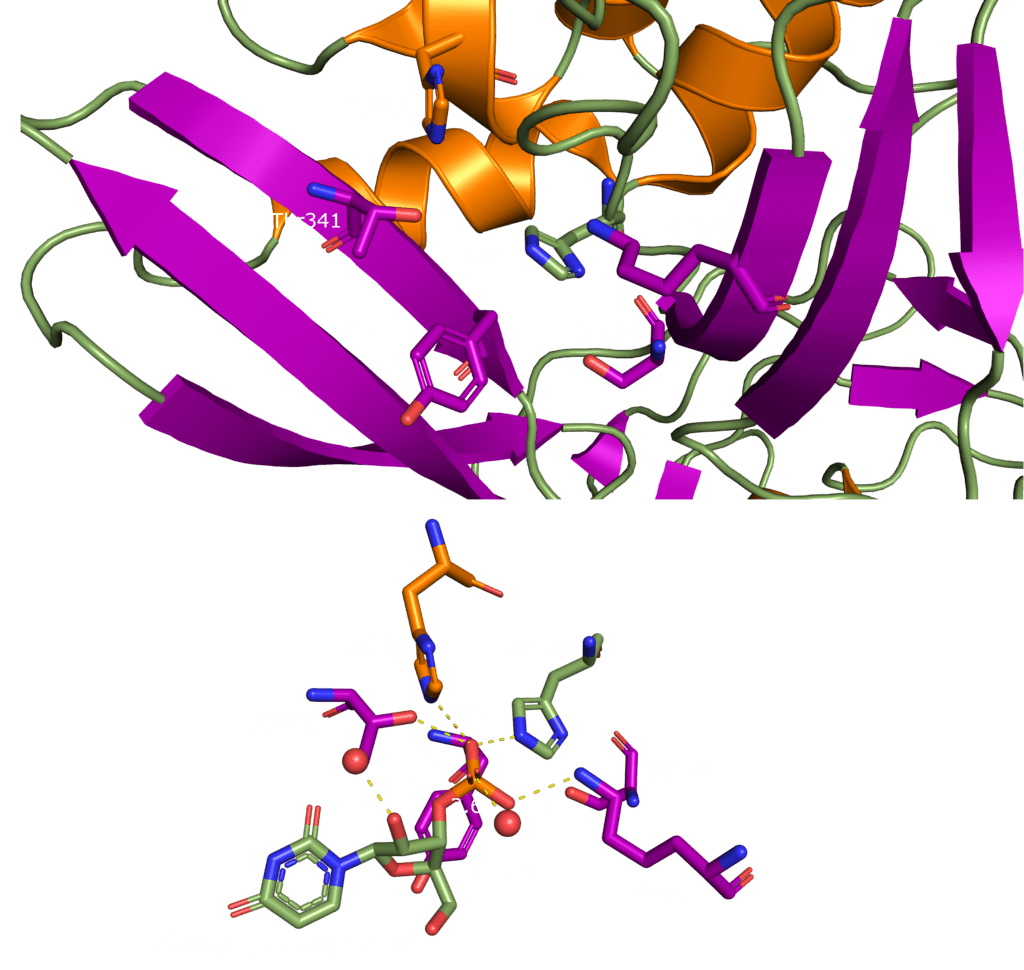
But What Does it do?
After all that we have a pretty clear picture of what Nsp15 NendoU looks like, but what does it actually do? The fact that it cuts RNA would immediately suggest a role in viral replication, but Nsp15 deficient coronaviruses are still able to replicate. So maybe not, at least it's not essential for replication. Another suggestion is that Nsp15 is involved in interfering with the hosts innate immune response, but other studies suggest this is independent of Nsp15 activity. Finally, it has been suggested that Nsp15 degrades viral RNA as a means of hiding viral infection from the host immune system. So why does coronavirus bother with Nsp15? I’m afraid we don’t exactly know yet, but we’re working on it.
With that I’m going to leave you with one final Voltron reference for making it to the end. Good job, you earned this.