During Christmas holidays in Germany the SARS-CoV-2 infections still increased dramatically. In Miesbach, a small town near Munich and close to the Alpes, where daily winter tourism was booming during Christmas, the hospital of Agatharied exceeded the alarming value of Corona infections. In December, 50 new infections occurred among patients and staff from several different wards all over the hospital.
As one of the chief doctors explained, the hospital had a great problem with patients being tested negative when arriving, but after a few days in the hospital being tested positive. 25 of these cases occurred in December. Either these patients got infected outside of the hospital a few days before arriving and were still in the incubation phase, when antigen concentrations are too low to get a positive rapid antigen test result - or these patients got infected during hospitalisation by other patients or the staff, which is more likely the case [1].
The hospital reacted by introducing stricter measurements for visitors, limiting the number of patients per room and also, and perhaps more importantly, by regularly testing the staff. Three test stations were set up and equipped with rapid antigen tests from Siemens and Roche. Two of the stations are mobile carriages that are moving from ward to ward testing the staff of the current shift and the third one is a fix station in a container room outside the building.
But during Christmas, staff capacities were already low while infections still increased, so new personnel was urgently required to test the doctors and the nursing, canteen and cleaning staff of the hospital. They were happy about every volunteer and even in Germany paperwork can be done pretty fast, so they managed to introduce daily shifts from 7:00 – 22:00h at all of the three test stations. Since our Corona Structural Task Force was on holiday, I had some time left to help out.

Testing Procedure
In a short time, the hospital of Agatharied had the capacities to test all ~1100 employees every day before their start of duty at one of the three test stations. All volunteers got a hygiene training and a quick instruction on how to use the two antigen test sets, how to register the tested staff and what to do in case of a positive test result.
To enable a fast communication of testing results and accelerate the registration process they introduced a bar code system with every employee having a personal code and their personal data deposited. 15 minutes after testing the result is entered into the system and an automatically generated email is sent to the tested person. If a positive test result occurs, the tested employee is called immediately to minimize his/her further contacts. Positive tested staff is subsequently tested by a PCR test in order to eliminate false positive test results.
In Agatharied we used the rapid antigen tests from Roche and Siemens. According to the Bundesinstitut für Arzneimittel und Medizinprodukte the Siemens test exhibits a sensitivity of 96.72% and a specificity of 99.22%. The Roche test exhibits values of 96.52% and 99.68% in sensitivity and specificity [2].
Here’s a quick overview on how to collect and prepare a sample for a Roche test [3]:
- Inserting a sterile swab into the patient’s nostril and swabbing the surface of the posterior nasopharynx.
- Inserting the swab into an extraction buffer tube and stir the swab a few times.
- Removing the swab while squeezing the sides of the tube to extract the sample from the swab.
- Pressing the nozzle cap onto the tube.
- Applying 3 drops of extracted sample to the specimen well of the test device.
- Reading the test result at 15 – 30 min.

Figure 2A: Required Material to prepare the Sample and do the Antigen Test. 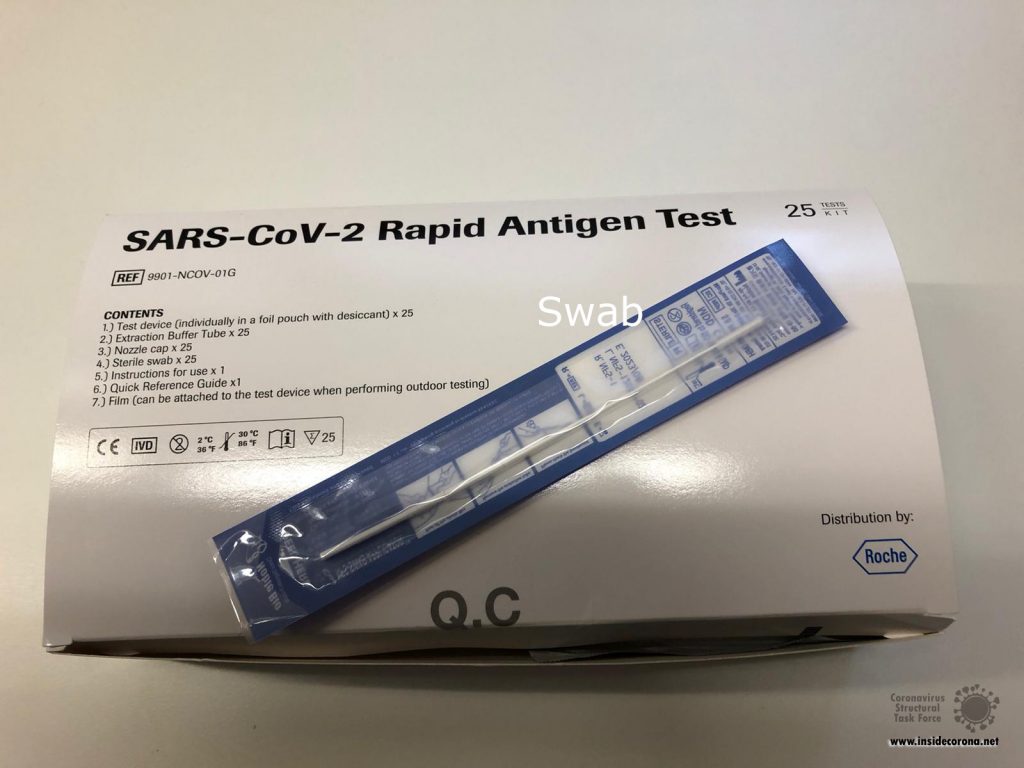
Figure 2B: Required Material for taking a Smear.

How do Rapid Antigen Tests work on molecular basis?
The posterior nasopharynx is where SARS-CoV-2 virions are highly concentrated. Concentrations can be 10 to 100 times higher than in the throat [4]. When the swab swabs the nasopharynx of an infected patient, SARS-CoV-2 virions are extracted. The extraction buffer then dissolves the viral envelope and the nucleocapsid protein is free. The nucleocapsid was chosen to be detected as an antigen, because it is highly conserved among all corona viruses and thus less likely to mutate, which is an important property considering that more and more mutations are spreading right now [5].
The test device is prepared with antibodies against the nucleocapsid antigen bound to molecules which form an immunoconjugate and cause a colour reaction. If the buffer solution contains nucleocapsid antigens, the test device shows a coloured test line indicating that the test is positive and the patient is infected. A second control line reacts with the immunoconjugate itself without any antigens bound to it. Hence, this line appears independently from the presence of antigens and proofs if the test was valid. [6]
The SARS-CoV-2 rapid antigen test only detects infected people in their most infectious phase, because of its higher detection limit compared to a PCR test. In this phase patients have a high virus concentration and are highly infectious. Nevertheless, negative tested patients can still be in the incubation phase before an infection or in the phase after an infection, where virus concentrations are too low to be detected by antigen tests. [7]
Thus, the rapid Antigen test is an effective way to filter a great amount of highly infectious SARS-CoV-2 patients in a short period of time. Nevertheless, a PCR test is still more reliable and should be done as a confirmation after a positive rapid Antigen test.
Statistics for December 2020
In December 3300 tests were done among the staff, of which 27 had a positive result confirmed by PCR. This makes a total rate of 2.6%. Since the introduction of the test stations after Christmas approximately 300 tests are done every day. [1]
References
[1] S. Grauvogl, Corona-Ausbruch in Krankenhaus: Mehr als 50 Neuinfektionen - auch Personal betroffen, Merkur.de, Published 30.12.21, Accessed 01.03.21, https://www.merkur.de/lokales/region-miesbach/hausham-ort74880/corona-bayern-ausbruch-krankenhaus-diffus-lage-kontrolle-agatharied-miesbach-90154653.html
[2] Bundesinstitut für Arzneimittel und Medizinprodukte, Liste der Antigen-Tests zum direkten Erregernachweis des Coronavirus SARS-CoV-2, Accessed 01.03.21, https://antigentest.bfarm.de/ords/f?p=101:100:22746556040321:::::&tz=1:00
[3] Diagnostics Roche, SARS-CoV-2 Rapid Antigen Test Quick Reference Guide.
[4] N. Ahsen, COVID-19-Tests: Nasopharynx-Abstrich, Accessed 01.03.21, Dtsch Arztebl 2020; 117(17): A-892 / B-748, https://www.aerzteblatt.de/archiv/213703/COVID-19-Tests-Nasopharynx-Abstrich
[5] S. Srinivasan et. al, Structural Genomics of SARS-CoV-2 Indicates Evolutionary Conserved Functional Regions of Viral Proteins, Viruses, 2020 Apr; 12(4): 360, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7232164/
[6] J. Lee et. al., A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2), tps://pubmed.ncbi.nlm.nih.gov/33099241/
[7] Diagnostics Roche, Infiziert gleich infektiös? Der diagnostische Unterschied zwischen dem SARS-CoV-2 Rapid Antigen Test und der PCR, Accessed 01.01.21, https://assets.cwp.roche.com/f/94122/x/caab12fe70/05-flyer-sars-cov-2_infiziert_v2_ansicht.pdf
