Scientific Illustrations
alternate image:
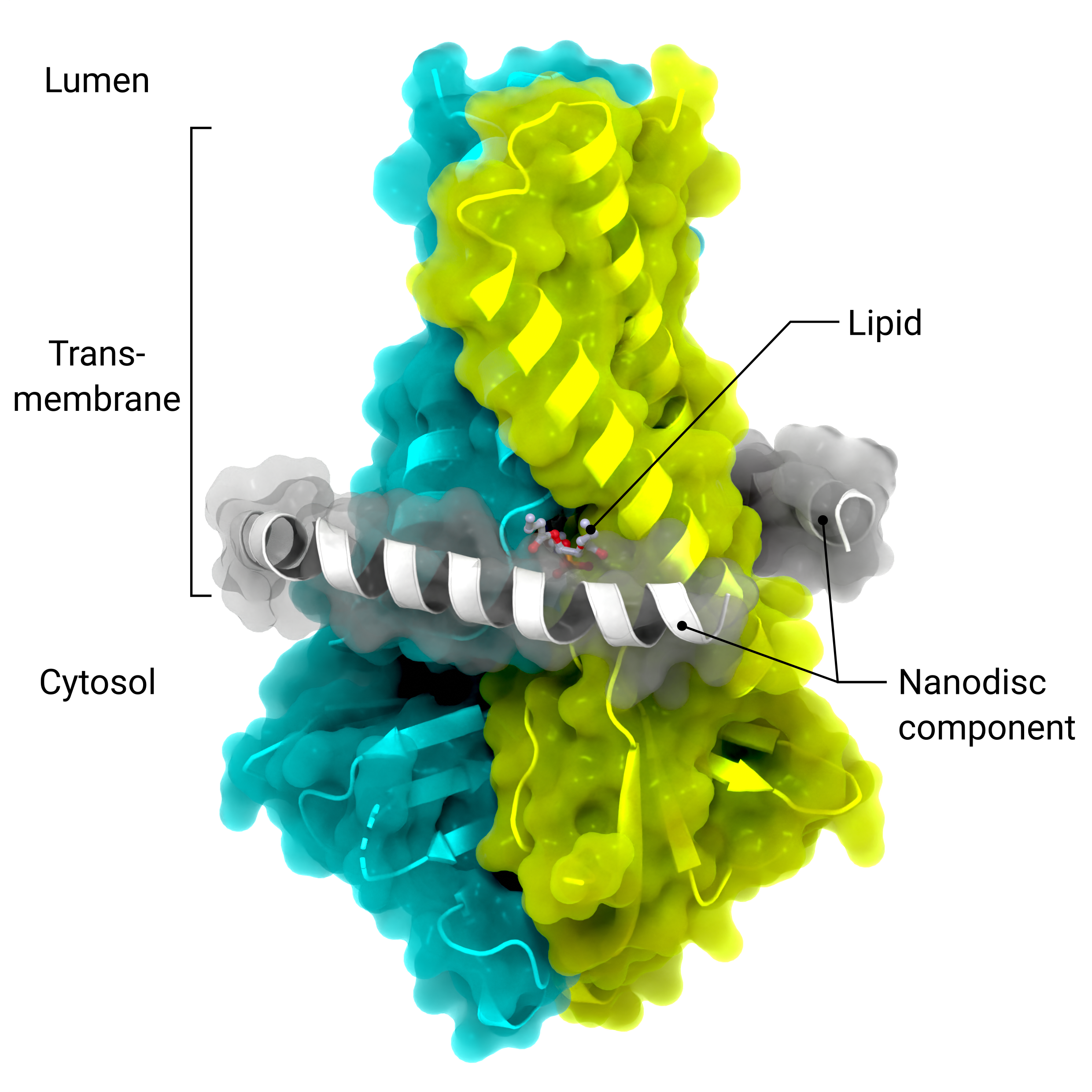
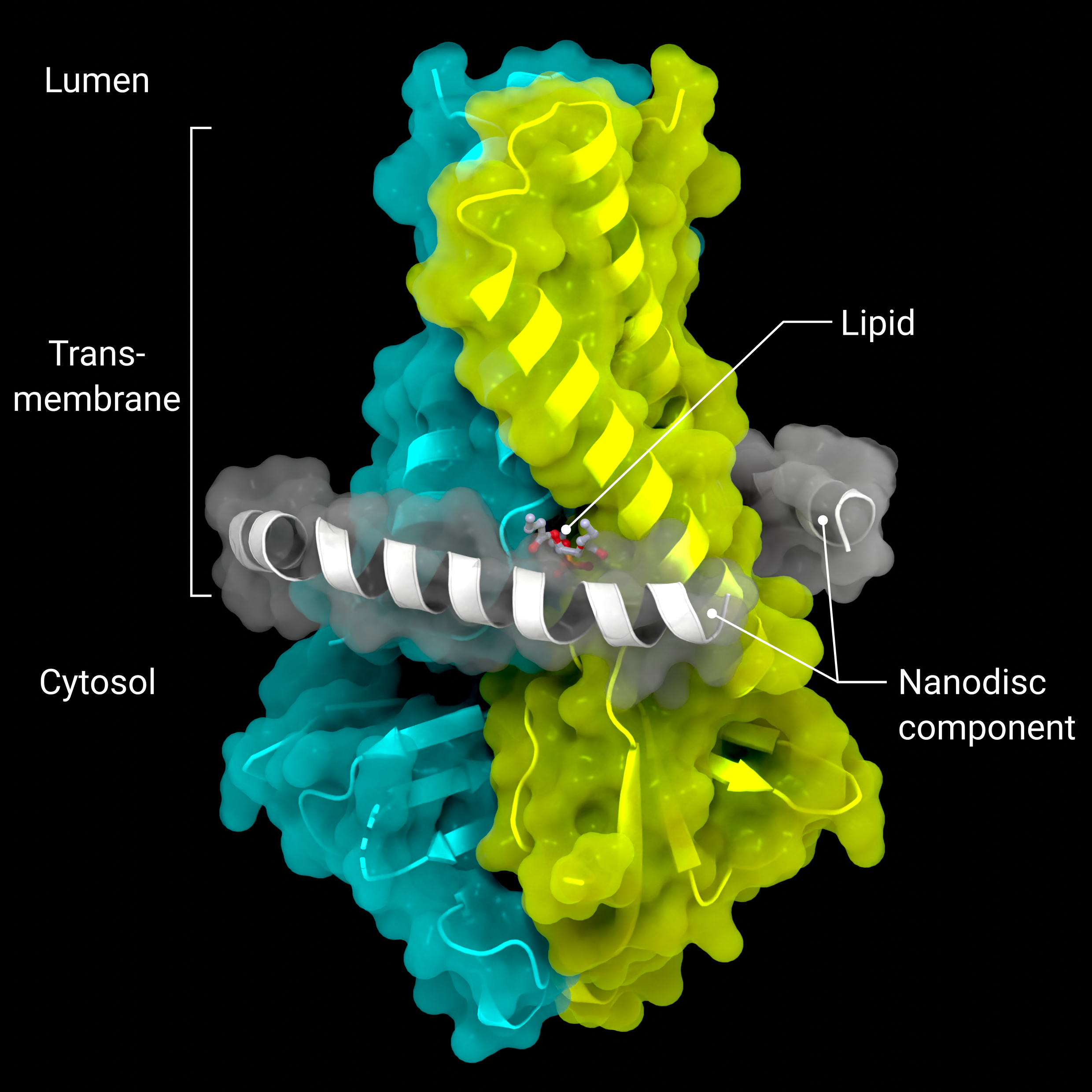
Accessory Protein ORF3a
The cryo-EM structure of the viriporin ORF3a (PDB: 7KJR) in a lipid nanodisc (ordered nanodisc components in grey)
Creator: Coronavirus Structural Task Force - Lisa Schmidt, David C. Briggs
License: cc-by-sa
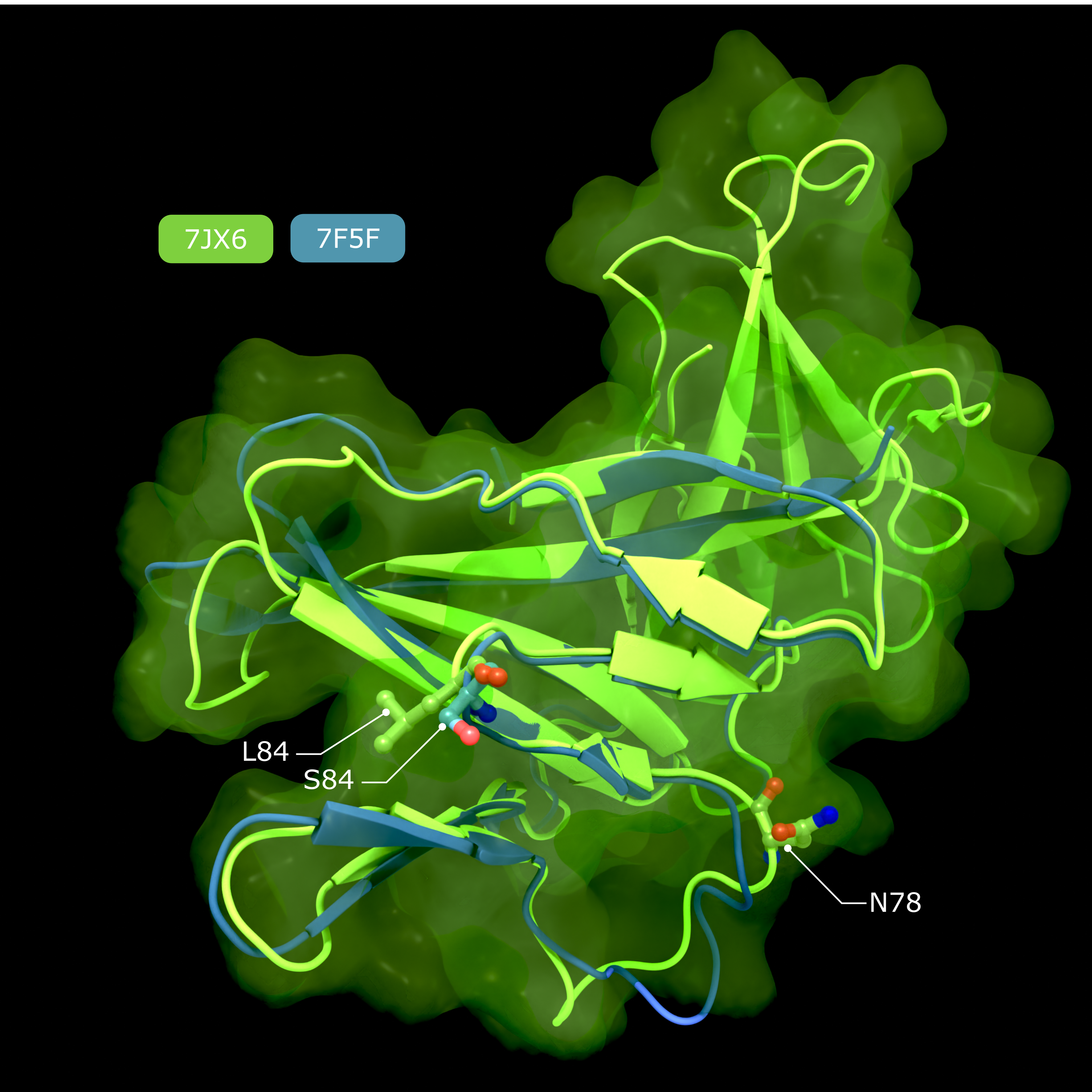
alternate image:
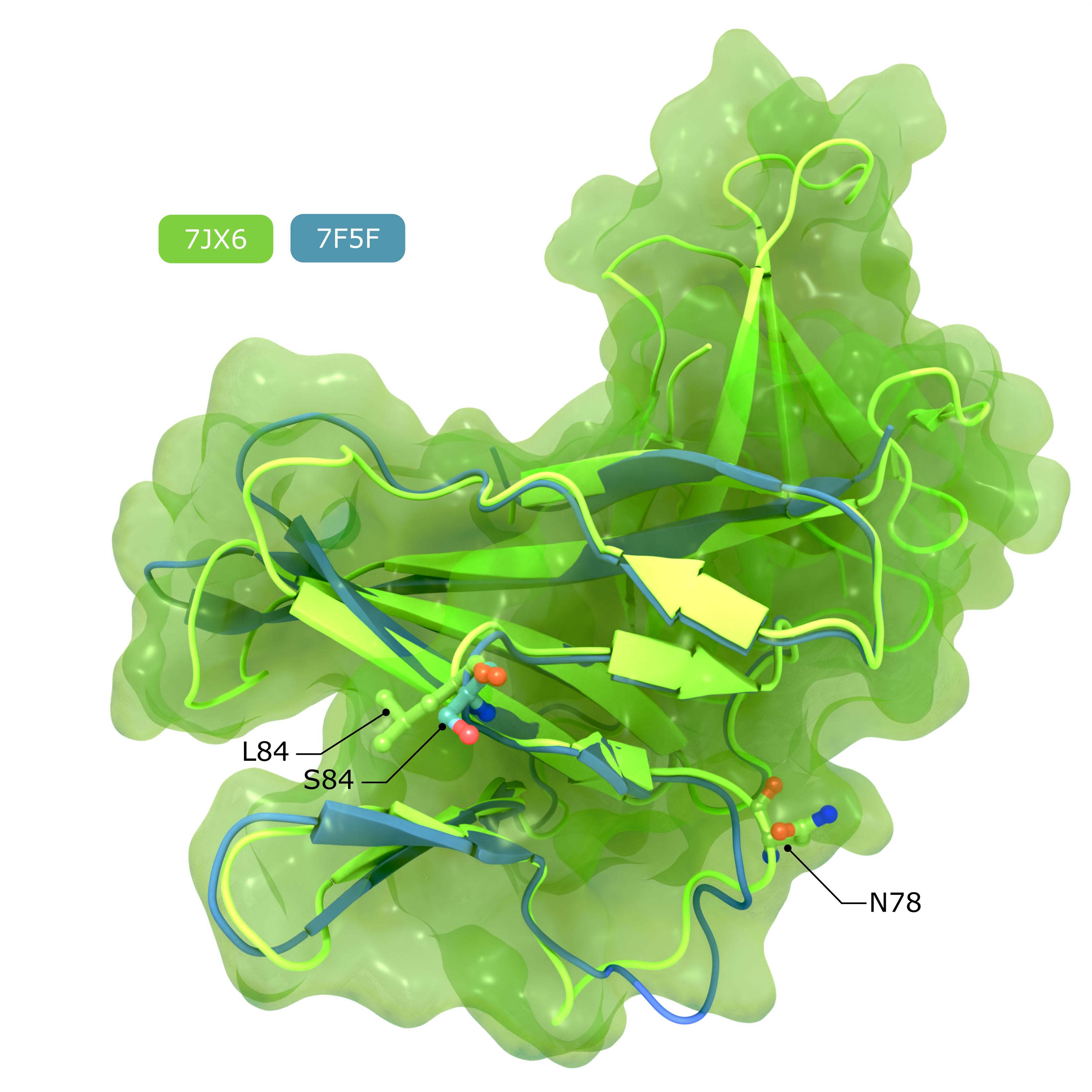
Accessory Protein ORF8
Comparison of structures of SARS-CoV-2 ORF8, showing the putative IL17RA binding surface that harbours the polymorphism at position 84, and the N-linked glycan that inhibits IL17RA binding in conventionally secreted ORF8.
Creator: Coronavirus Structural Task Force - Lisa Schmidt, David C. Briggs
License: cc-by-sa
alternate image:
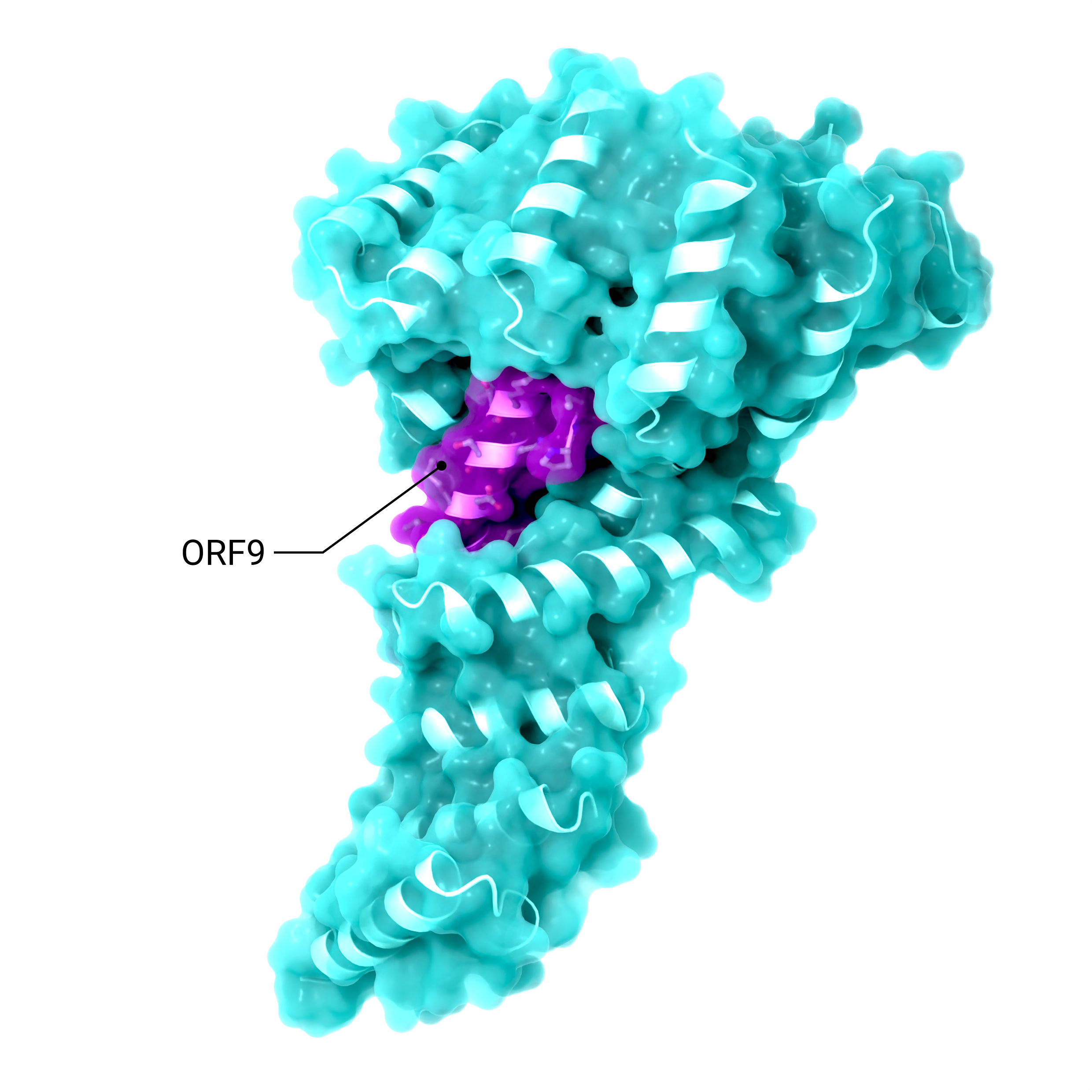
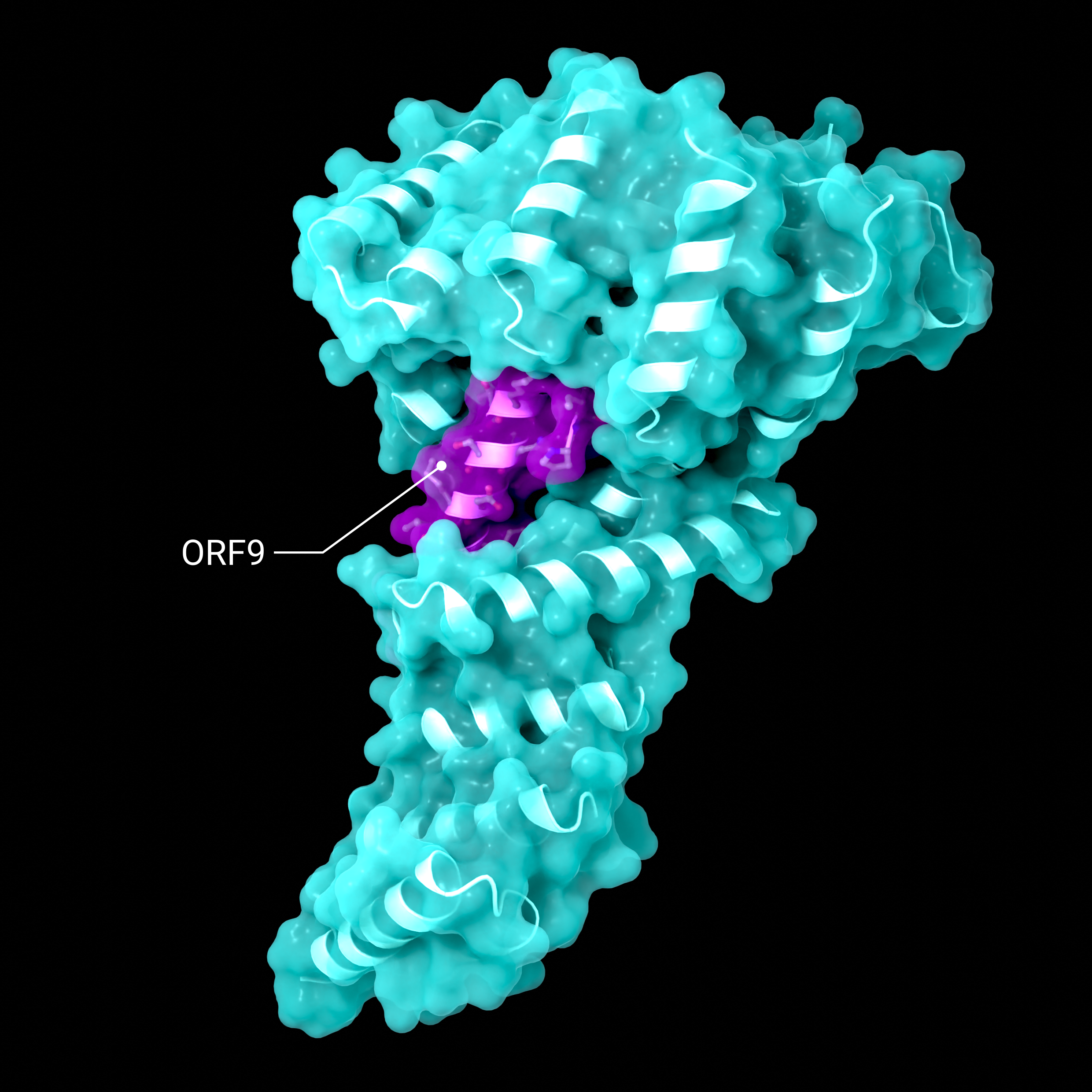
Accessory Protein ORF9
Structure showing the binding of SARS-CoV-2 ORF9 binding to the human mitochondrial transport proteins, TOM70
Creator: Coronavirus Structural Task Force - Lisa Schmidt, David C. Briggs
License: cc-by-sa
alternate image:
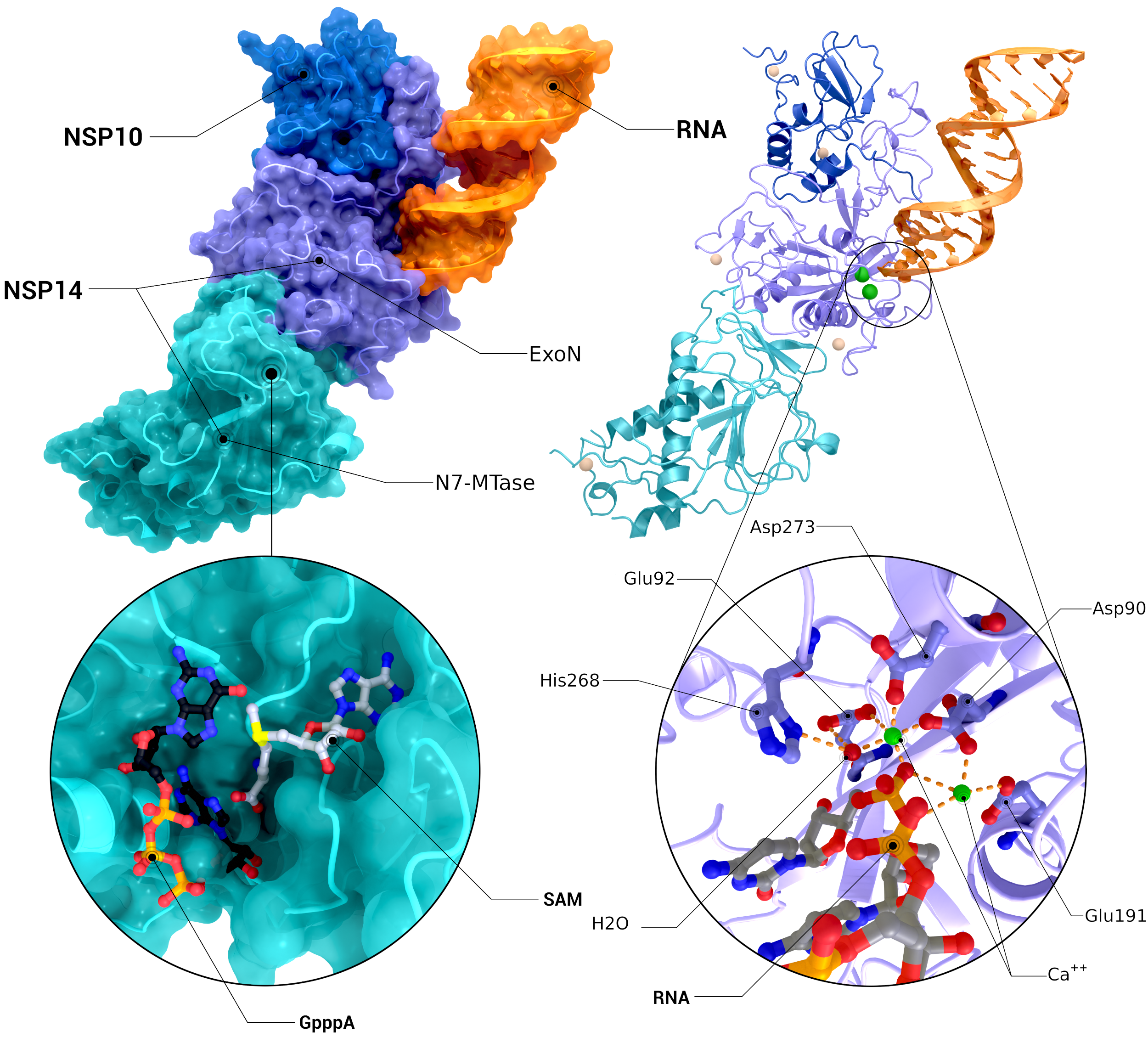
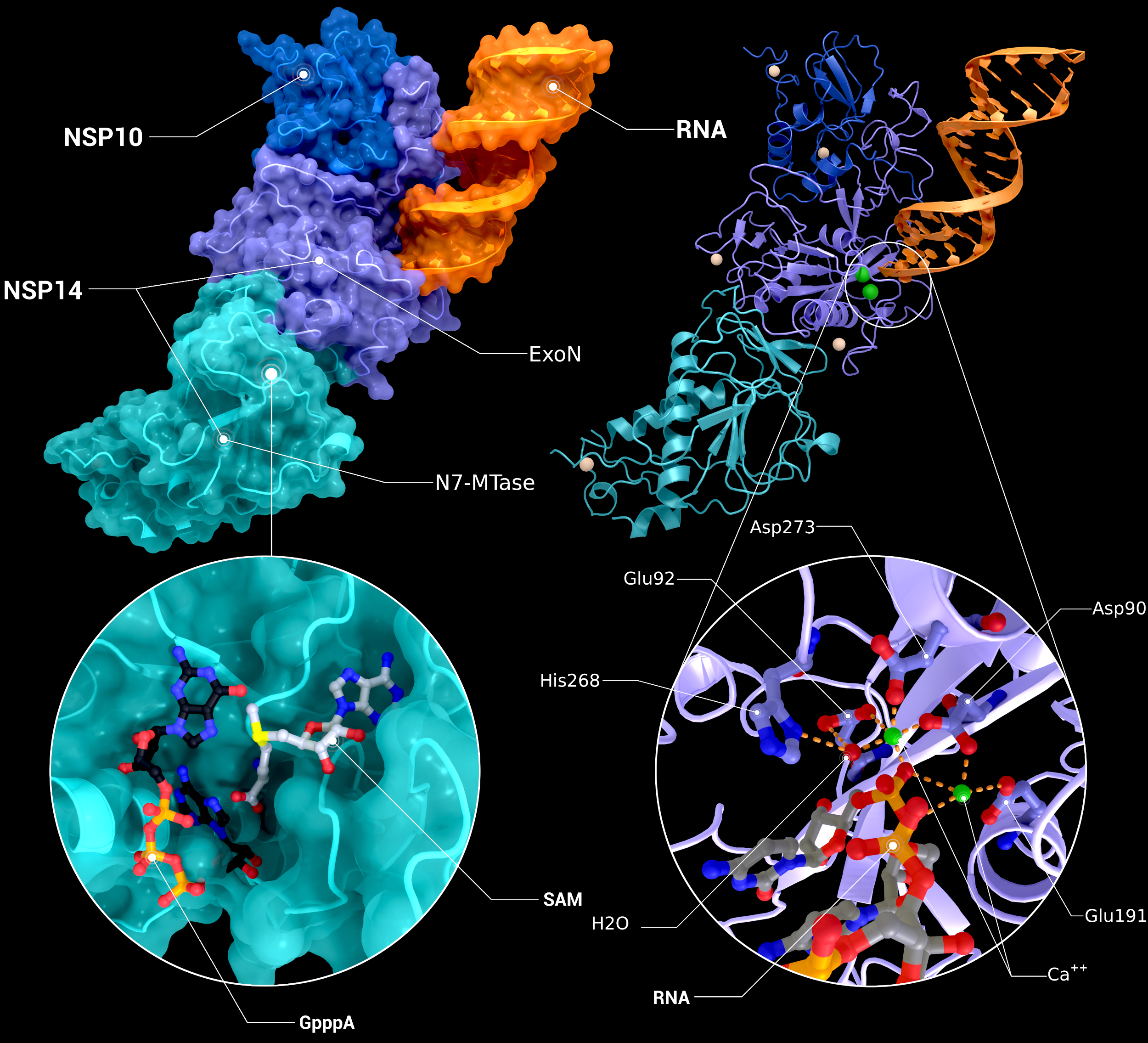
NSP10, NSP14 Heterodimer
The heterodimer of nsp10 with nsp14 bound to mismatched RNA bound in its Exoribonuclease domain (PDB ID: 7n0b(nsp10/14/RNA), 5c8s, 5c8t(N7-MTase introspection), 7n0b(ExoN introspection)).
Creator: Coronavirus Structural Task Force - Protein Imager, Cameron Fyfe
License: cc-by-sa
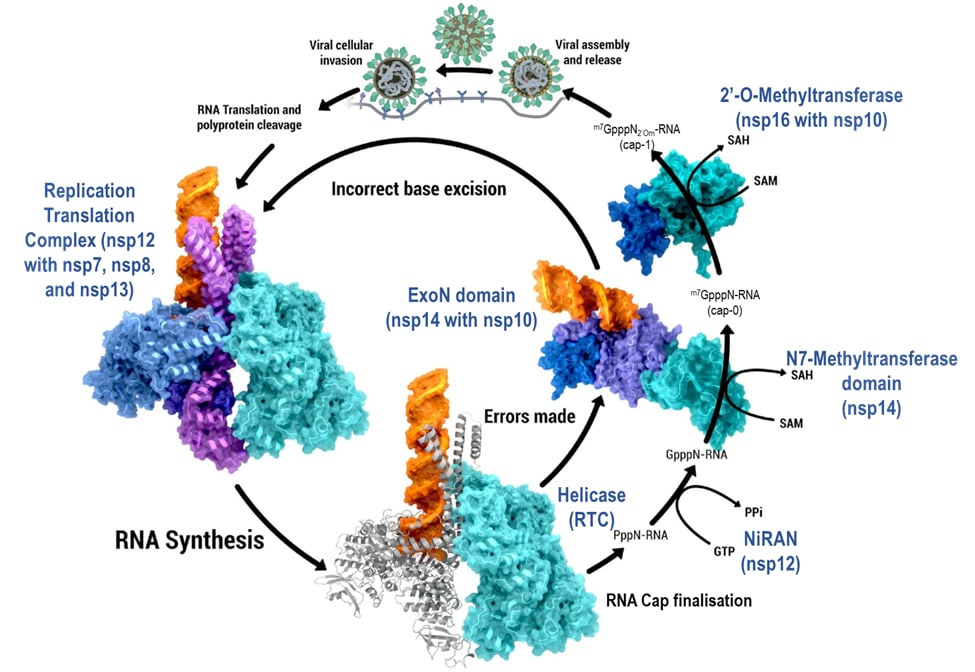
NSP10, NSP14, NSP16 - RNA Synthesis
RNA synthesis, fidelity regulation and 5’ RNA cap finalization. Processes, products and steps within viral reproduction and RNA synthesis shown in black text; proteins and protein complexes performing each step shown in blue text (PDB ID: 6xez (RTC), 7n0b (nsp10/14/RNA), 7l6t (nsp10/16)).
Creator: Coronavirus Structural Task Force - Protein Imager, Cameron Fyfe
License: cc-by-sa
alternate image:
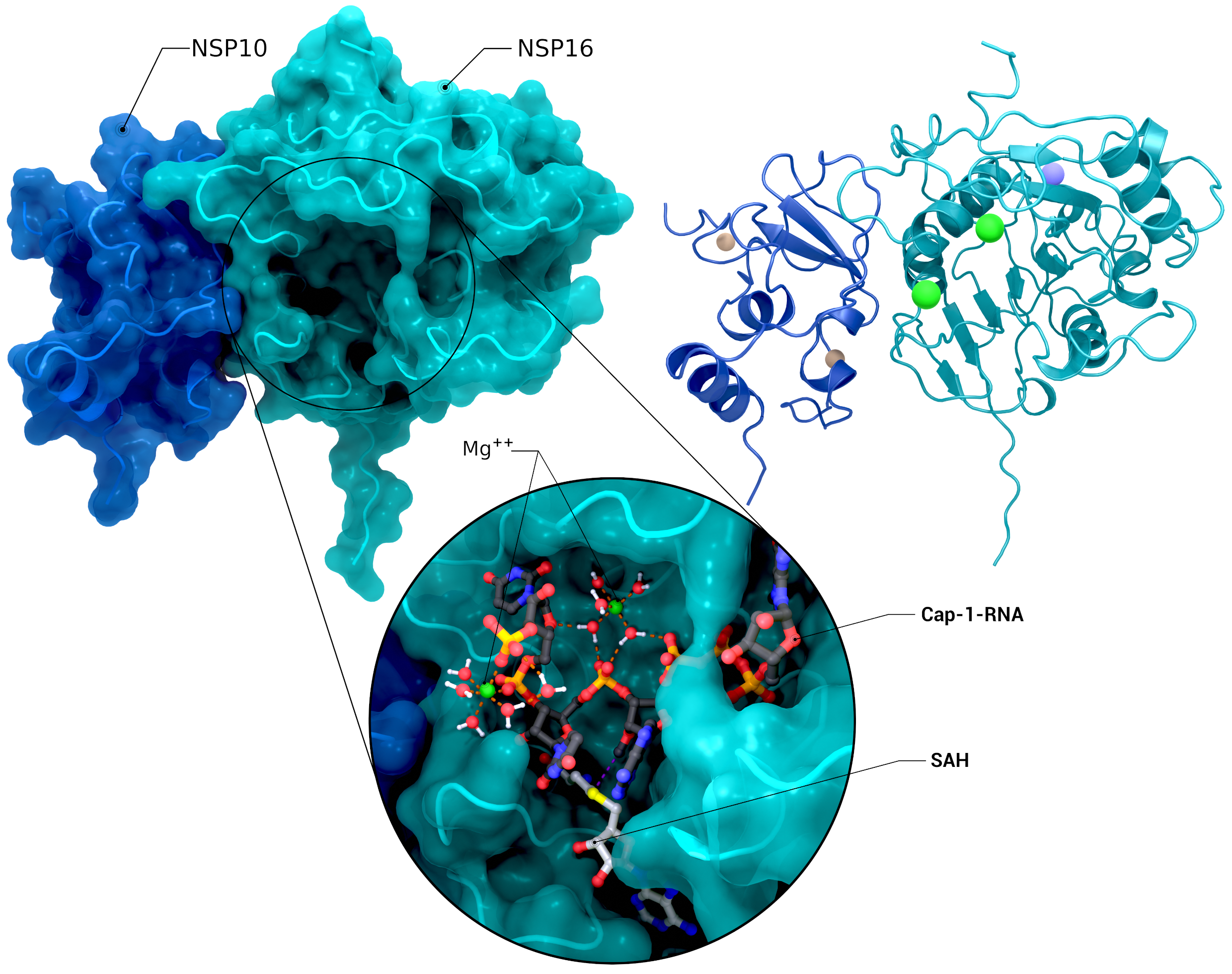
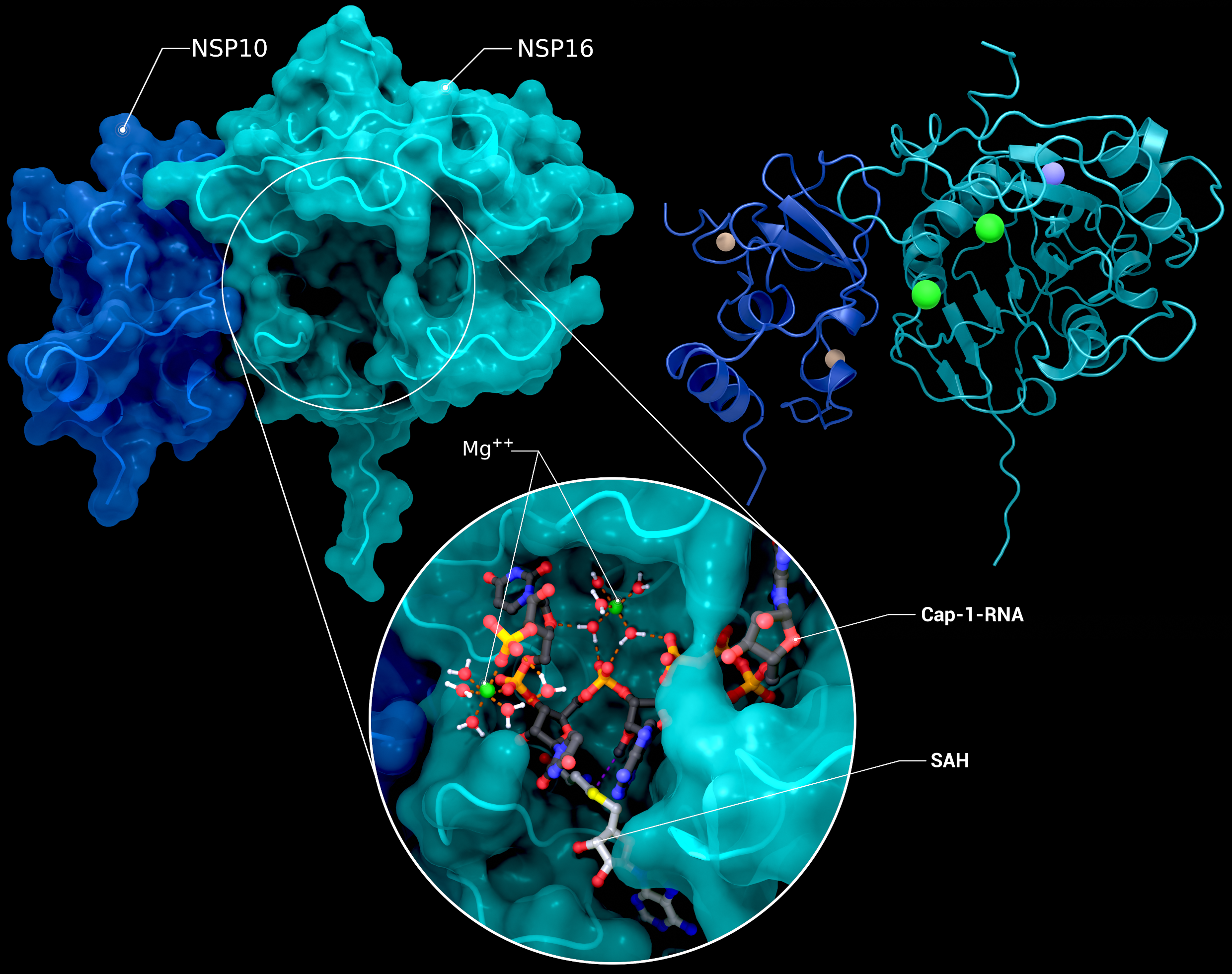
NSP10, NSP16 Heterodimer
The heterodimer of nsp10 with nsp16 in a postreaction state with magnesium, its Cap-1-RNA product, and S-adenosyl-L-homocysteine (PDB ID: 7l6t (nsp10/16), 7jyy (nsp16 2′-O-MTase introspection)).
Creator: Coronavirus Structural Task Force - Protein Imager, Cameron Fyfe
License: cc-by-sa
alternate image:
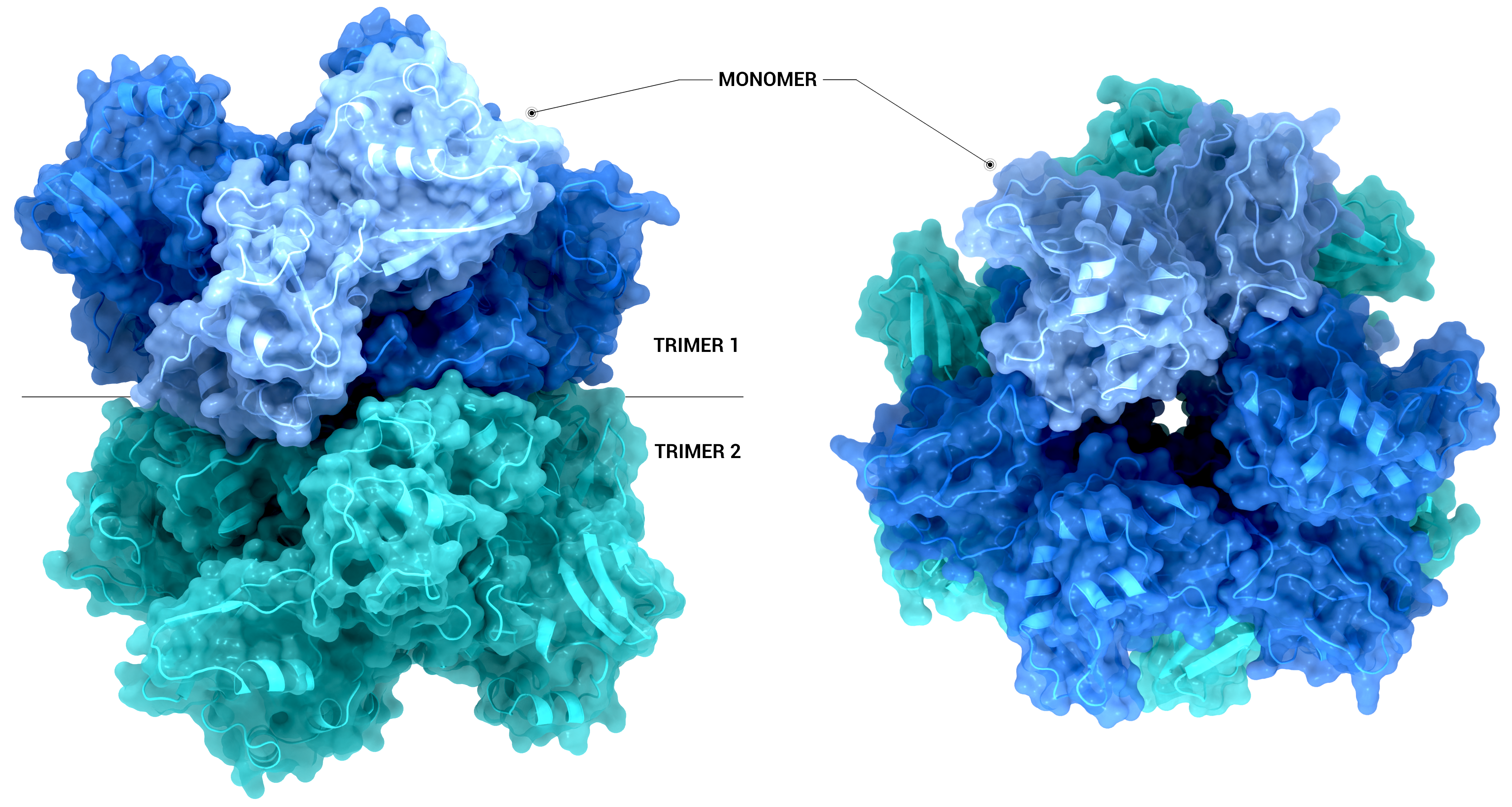
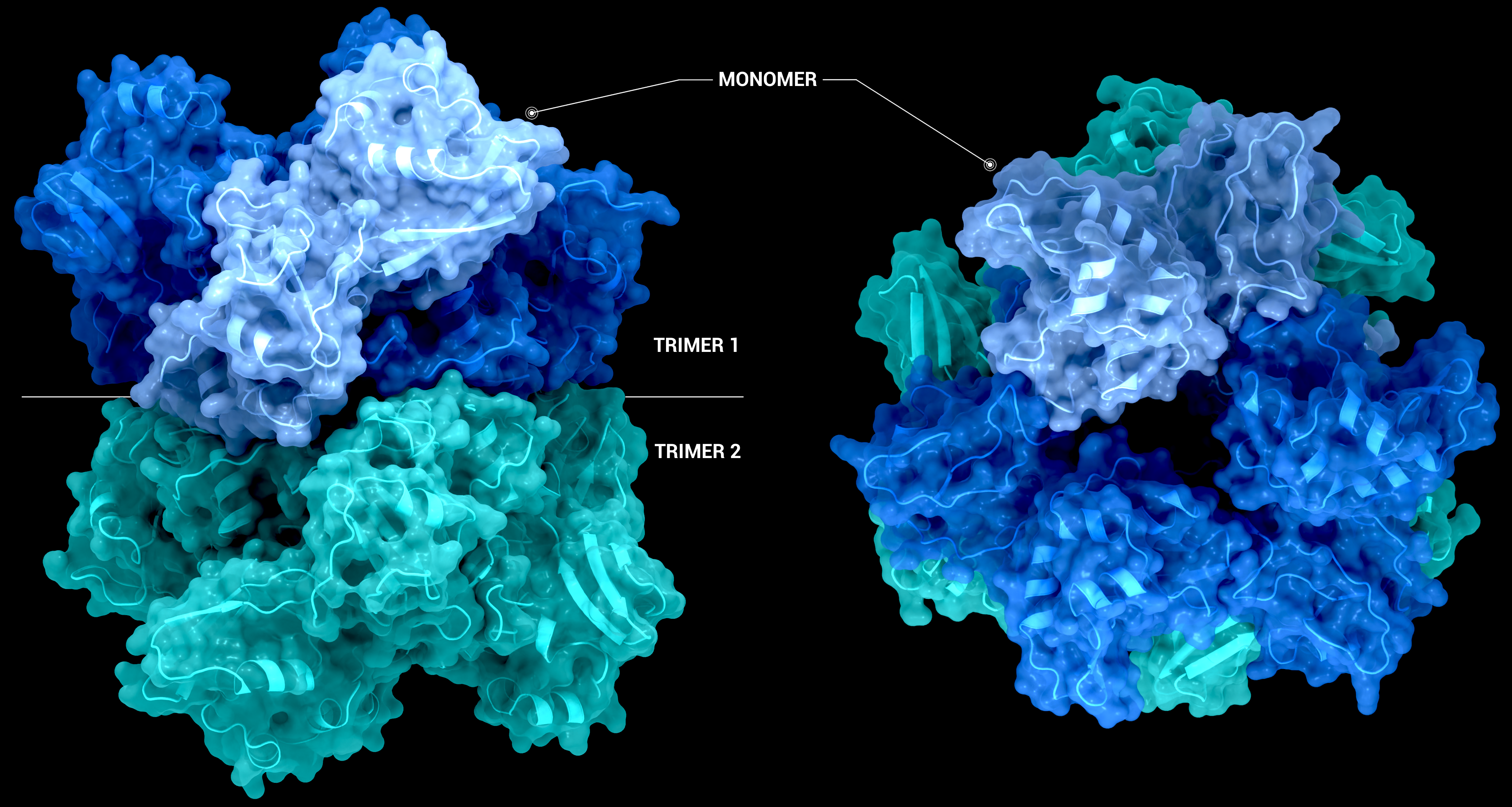
NSP15 Endoribonuclease NendoU - Hexamer
The Structure of the nsp15 hexamer generated by crystallographic symmetry using PDB entry 6X4I. On the left-hand side, the nsp15 hexamer is represented as a transparent surface and cartoon from a side-on view. On the right-hand side, the hexamer has been
Creator: Coronavirus Structural Task Force - Protein Imager, Sam Horrell
License: cc-by-sa
alternate image:
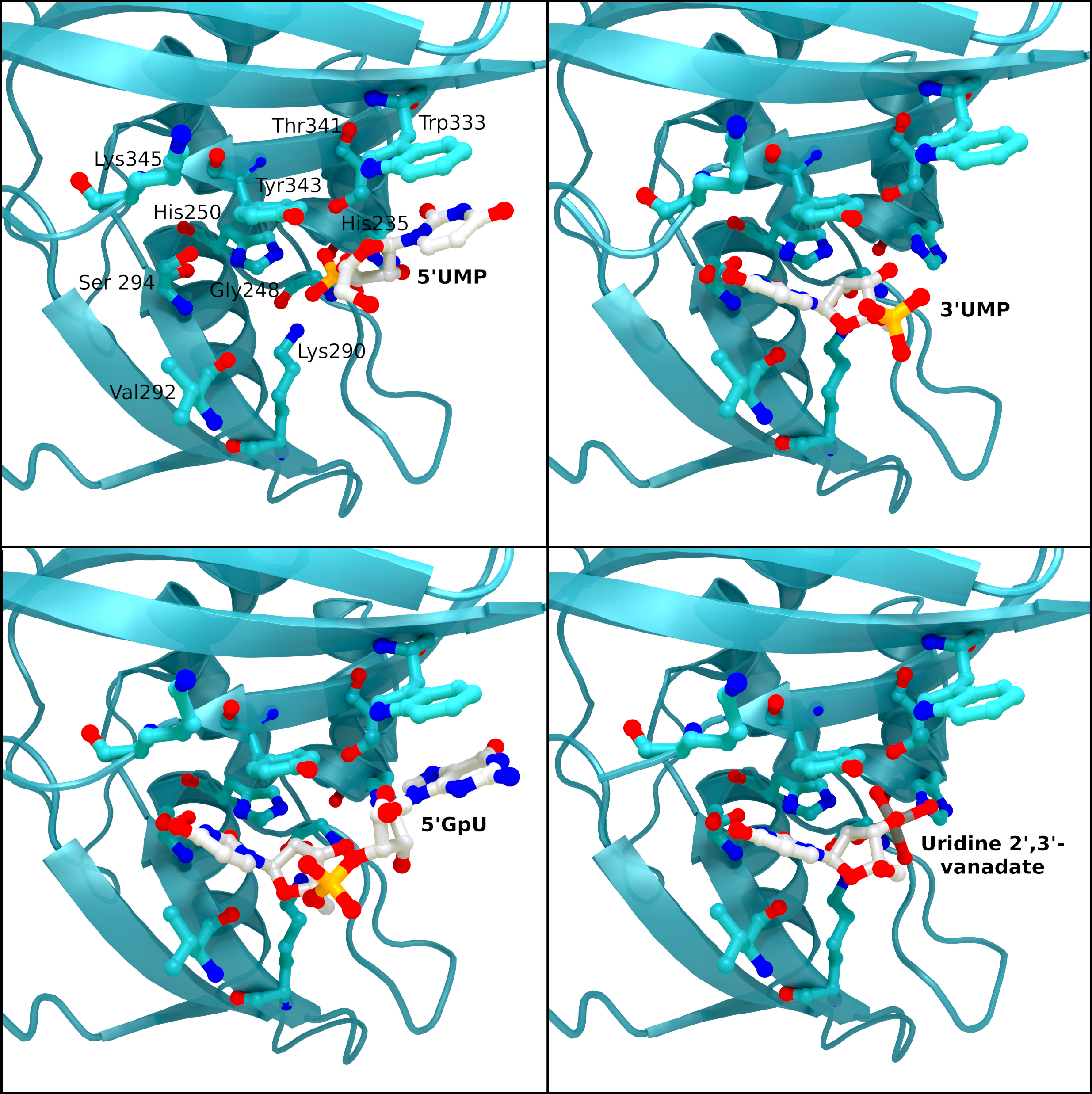
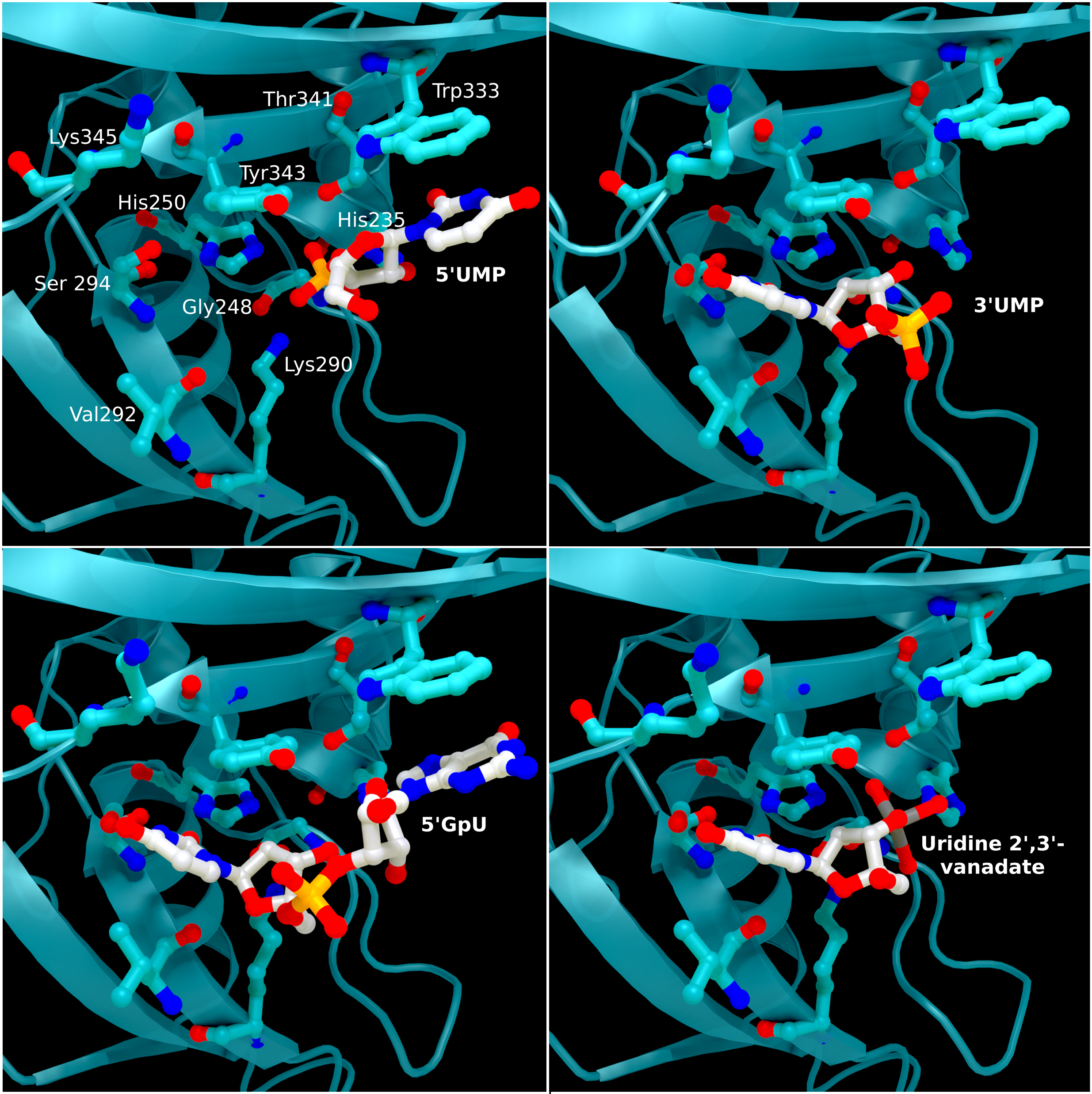
NSP15 Endoribonuclease NendoU - Ligands
SARS-CoV 2 nsp15 active site crystal structures with bound reaction intermediates. 5′UMP (PDB entry: 6WLC) in the top left, 3′UMP (PDB entry: 6X4I) in the top right, 5′GpU (PDB entry: 6X1B) in the bottom left, and the cyclic intermediate mimic uridine 2′,
Creator: Coronavirus Structural Task Force - Protein Imager, Sam Horrell
License: cc-by-sa

alternate image:
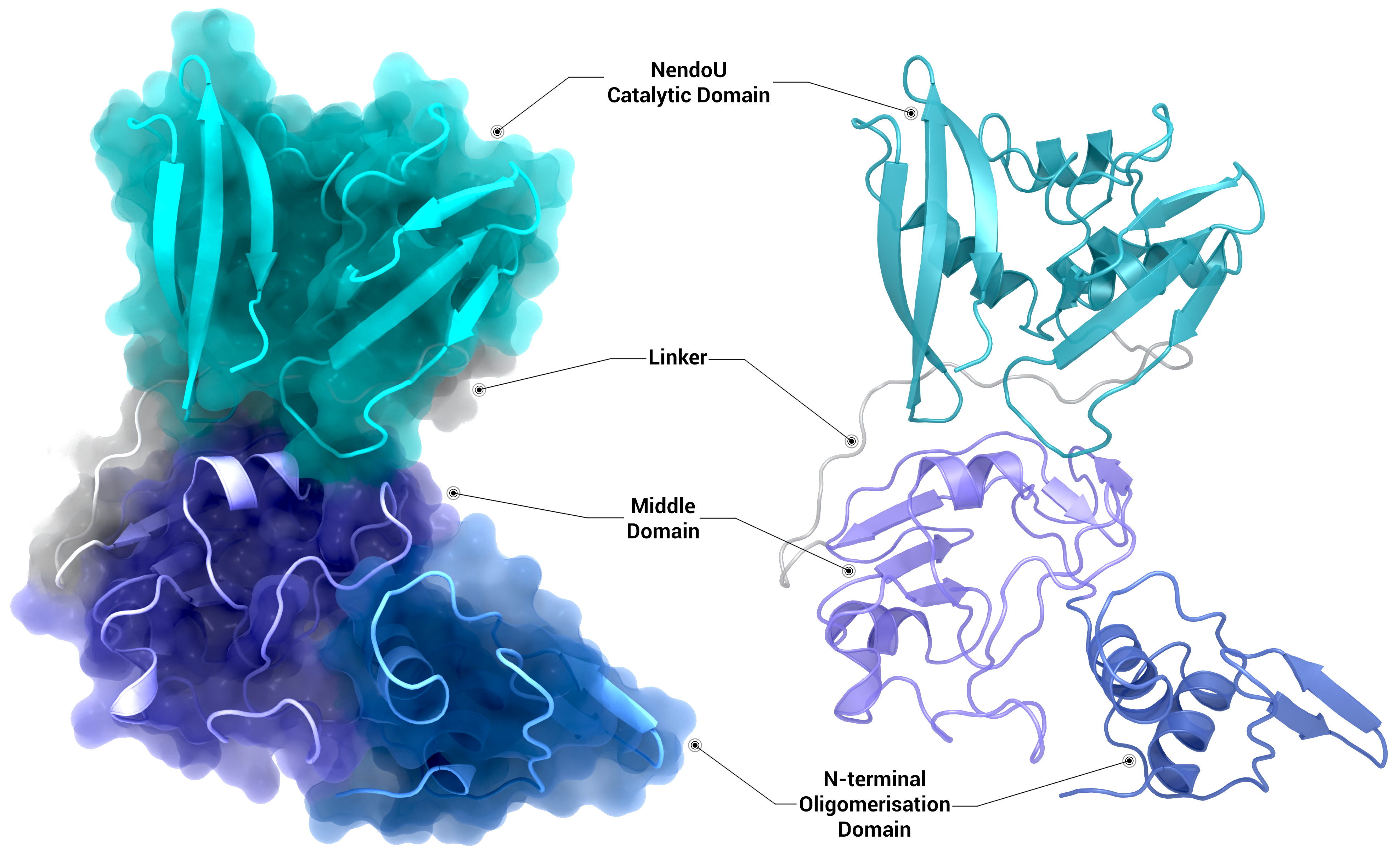
NSP15 Endoribonuclease NendoU - Monomer
Crystal structure of the nsp15 monomer represented as a transparent surface and cartoon (left) and as a cartoon (right) coloured by domain using PDB entry 6X4I.
Creator: Coronavirus Structural Task Force - Protein Imager, Sam Horrell
License: cc-by-sa
alternate image:
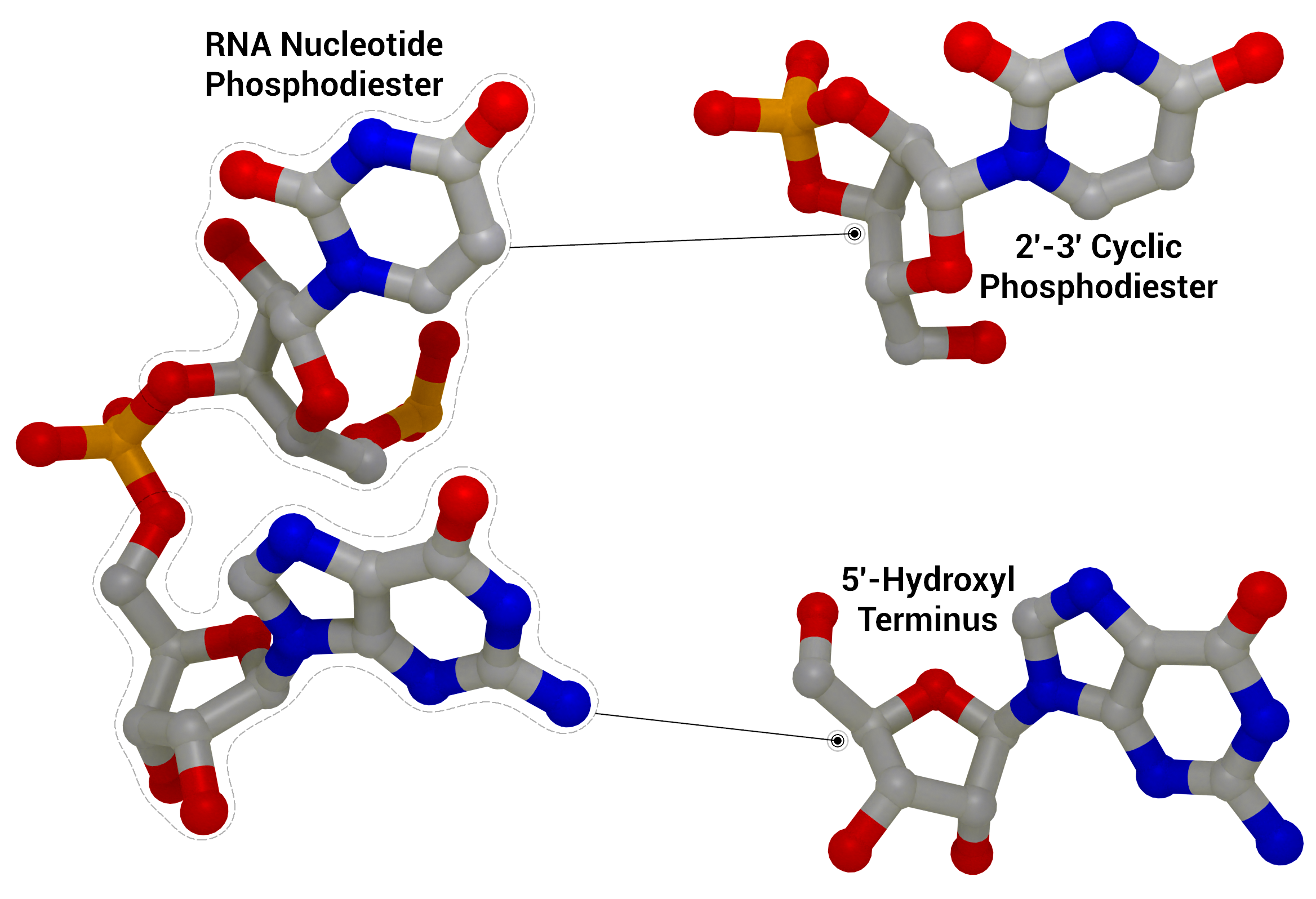
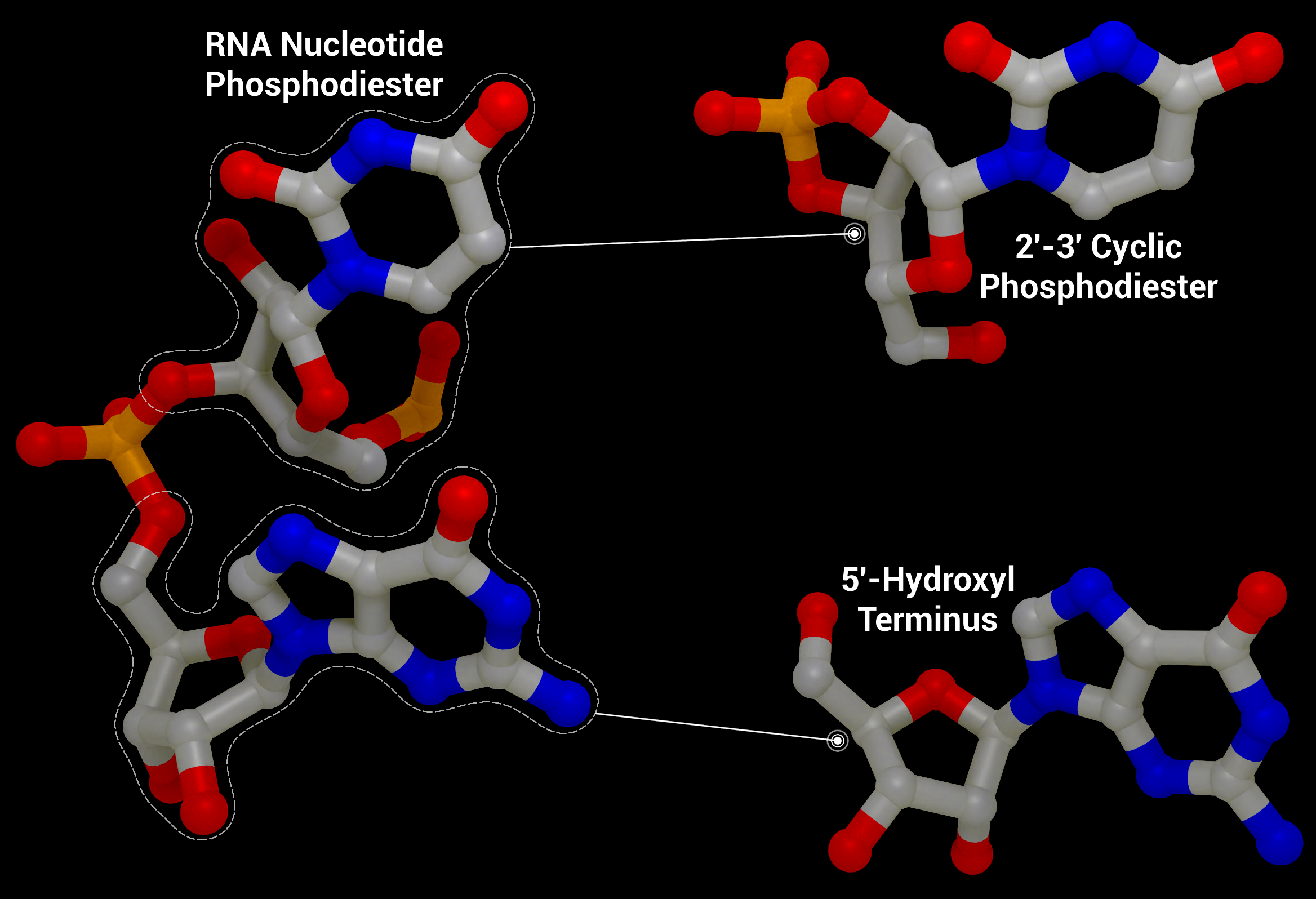
NSP15 Endoribonuclease NendoU - RNA Cleavage
RNA Cleavage as performed by nsp15 to give a 2′‐3′ cyclic phosphodiester and 5′‐hydroxyl terminus from an RNA nucleotide phosphodiester from PDB entry 1RNA.
Creator: Coronavirus Structural Task Force - Protein Imager, Sam Horrell
License: cc-by-sa
alternate image:
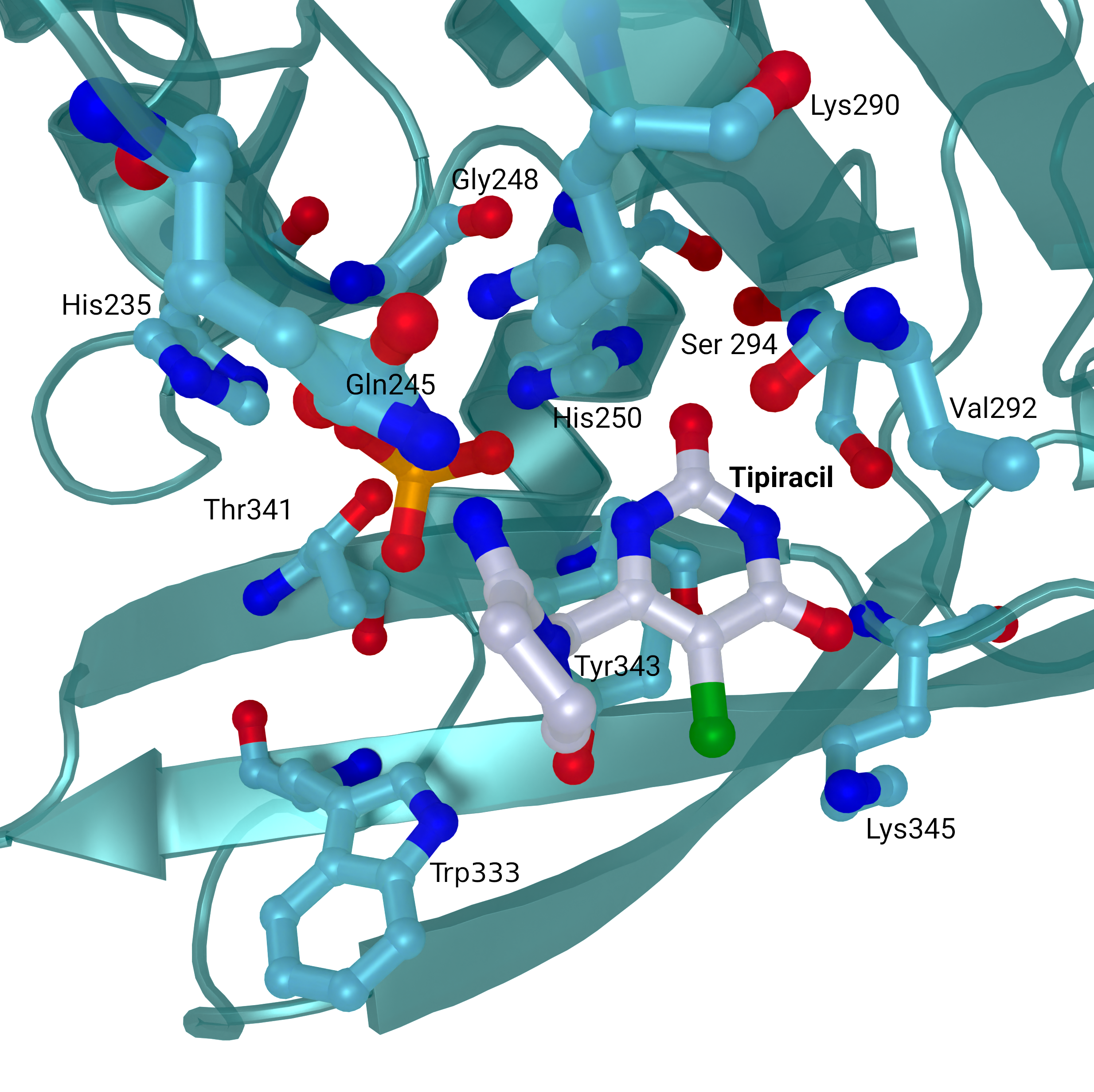
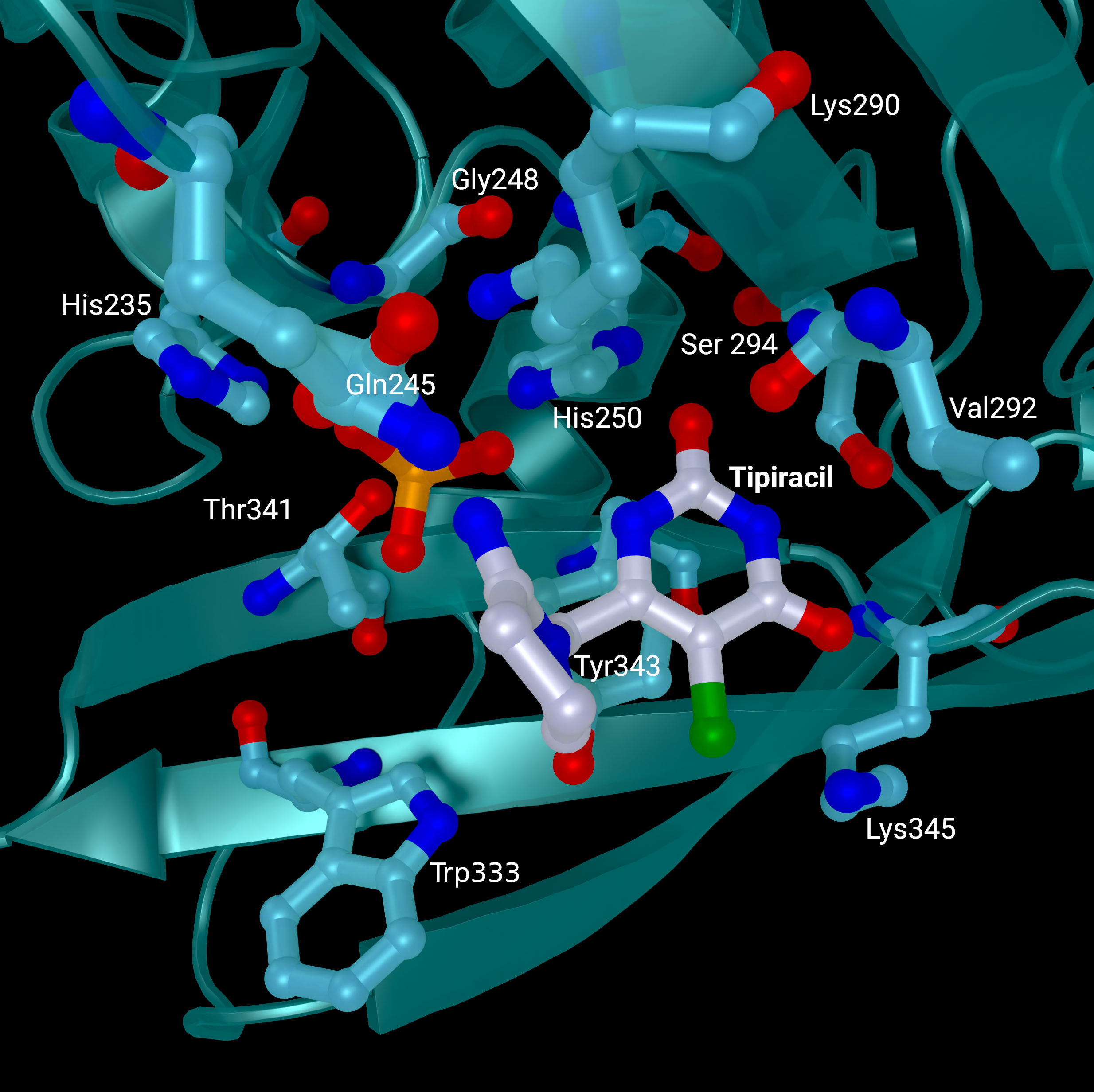
NSP15 Endoribonuclease NendoU - Tipiracil
SARS-CoV 2 nsp15 active site crystal structures with bound Tipiracil from PDB entry 6WXC. The protein is coloured in teal and represented as a cartoon with active site residues and bound Tipiracil represented as sticks. Tipiracil is coloured white.
Creator: Coronavirus Structural Task Force - Protein Imager, Sam Horrell
License: cc-by-sa
alternate image:
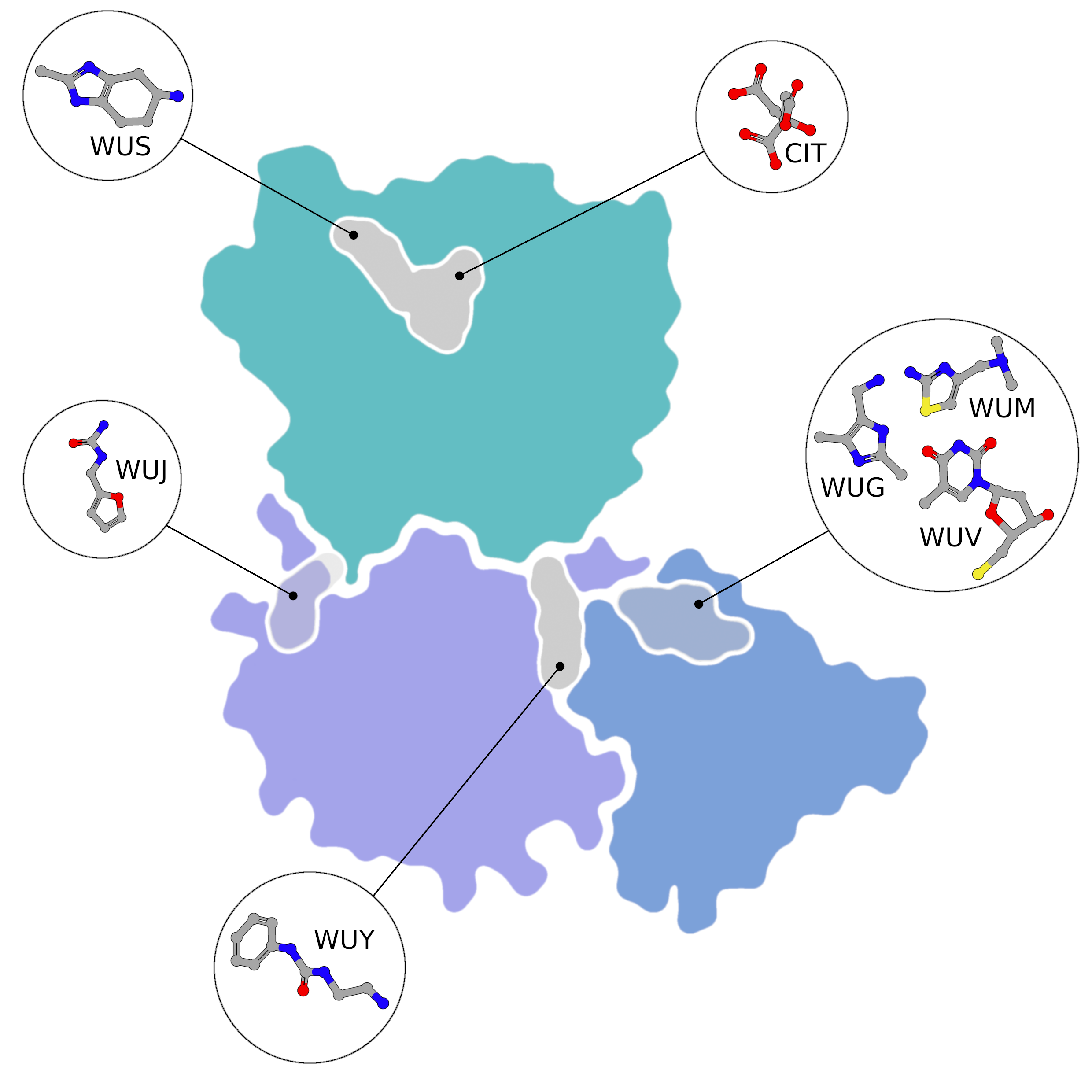
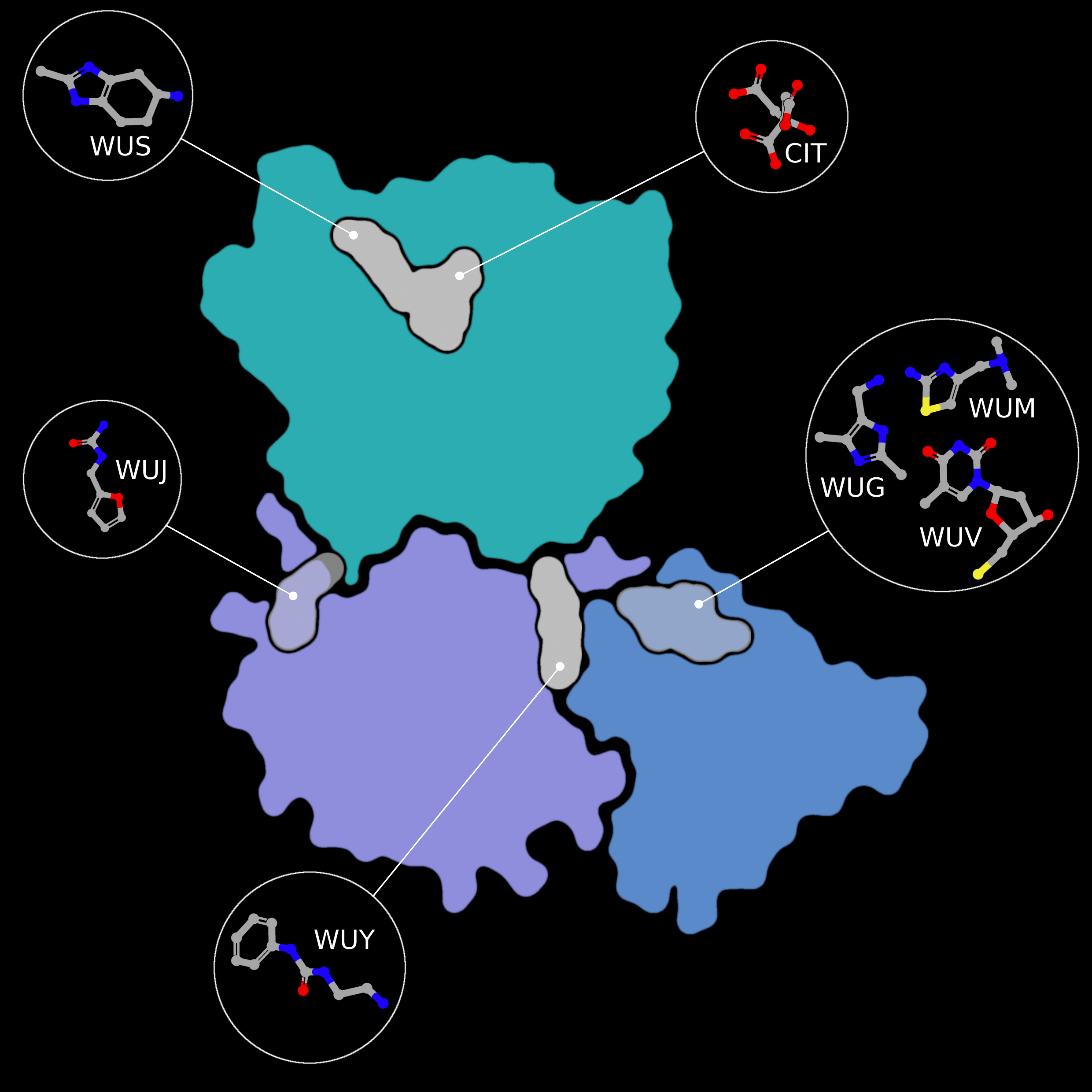
NSP15 Endoribonuclease NendoU - Topology
Small molecule fragment screening against SARS-CoV-2 nsp15, with nsp15 represented as flatfield coloured by domain (NendoU in teal, Middle Domain in purple, and N-terminal Oligomerisation domain in blue. Fragment binding is shown as a flat field, coloured
Creator: Coronavirus Structural Task Force - Protein Imager, Sam Horrell
License: cc-by-sa
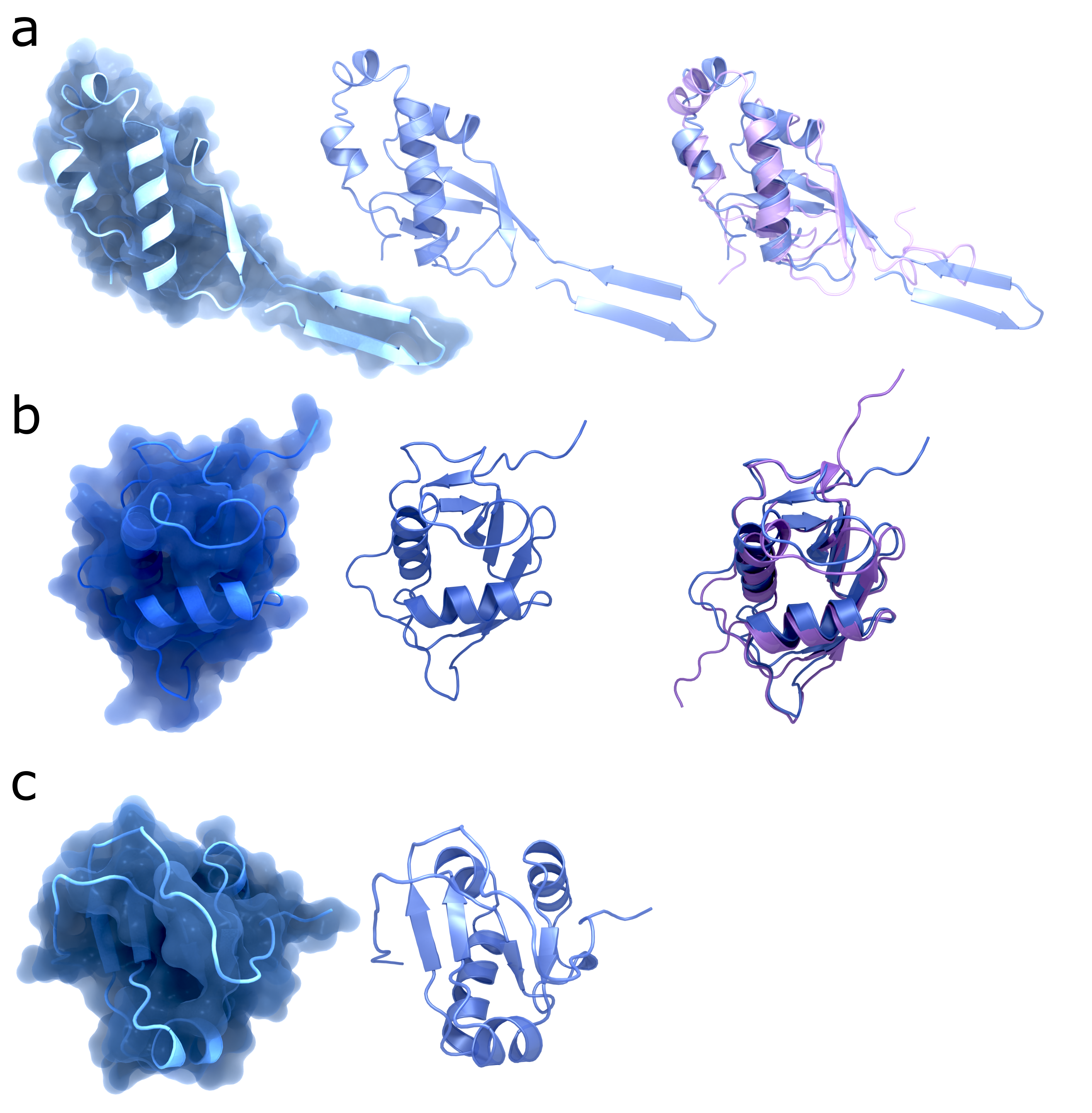
NSP3 Domain Comparison SARS-CoV-2 vs SARS-CoV-1
Structures from the NSP3 domains Ubl1 (a), NAB (b), and Y3 (c). The left column shows the structures of SARS-CoV-2 with surface (PDB codes: : 7KAG, 7LGO, 7RQG); the middle column shows the same structures without surface; the right column shows the struct
Creator: Coronavirus Structural Task Force - Protein Imager, Maximilian Edich
License: cc-by-sa
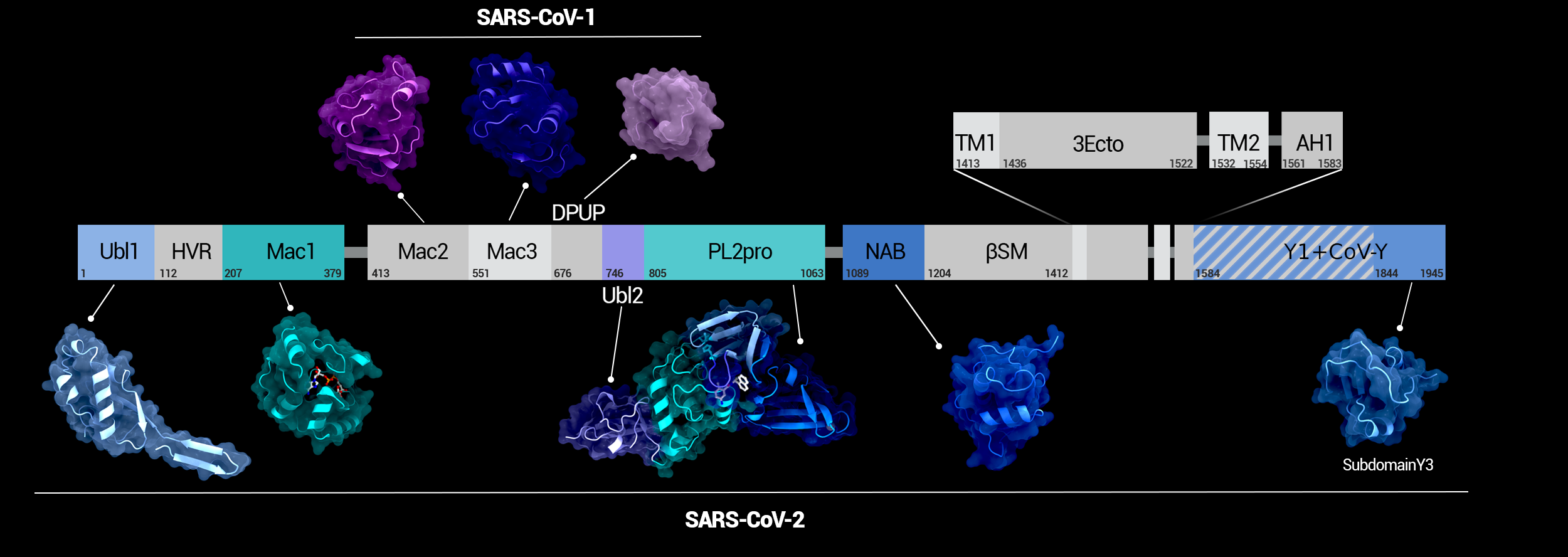
alternate image:
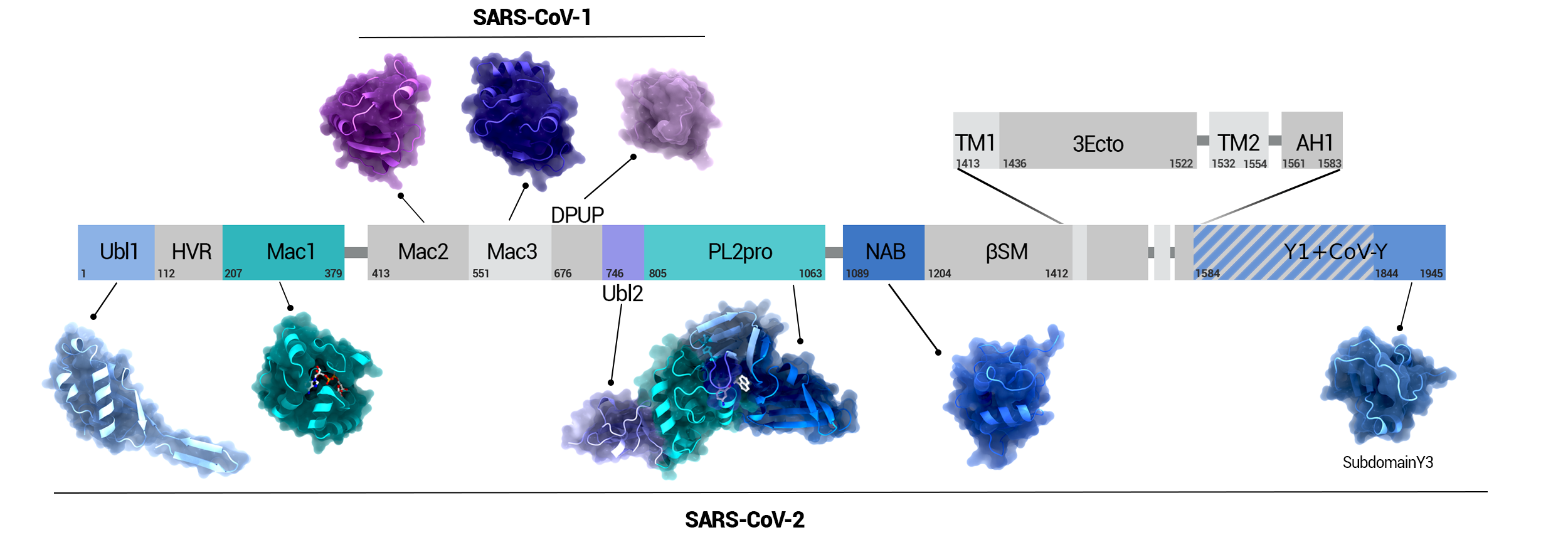
NSP3 Domain Overview
Overview of the domains of NSP3 and their position in the protein. Domains whose structure was solved in SARS-CoV-2 are highlighted in color.
Creator: Coronavirus Structural Task Force - Protein Imager, Maximilian Edich
License: cc-by-sa
alternate image:
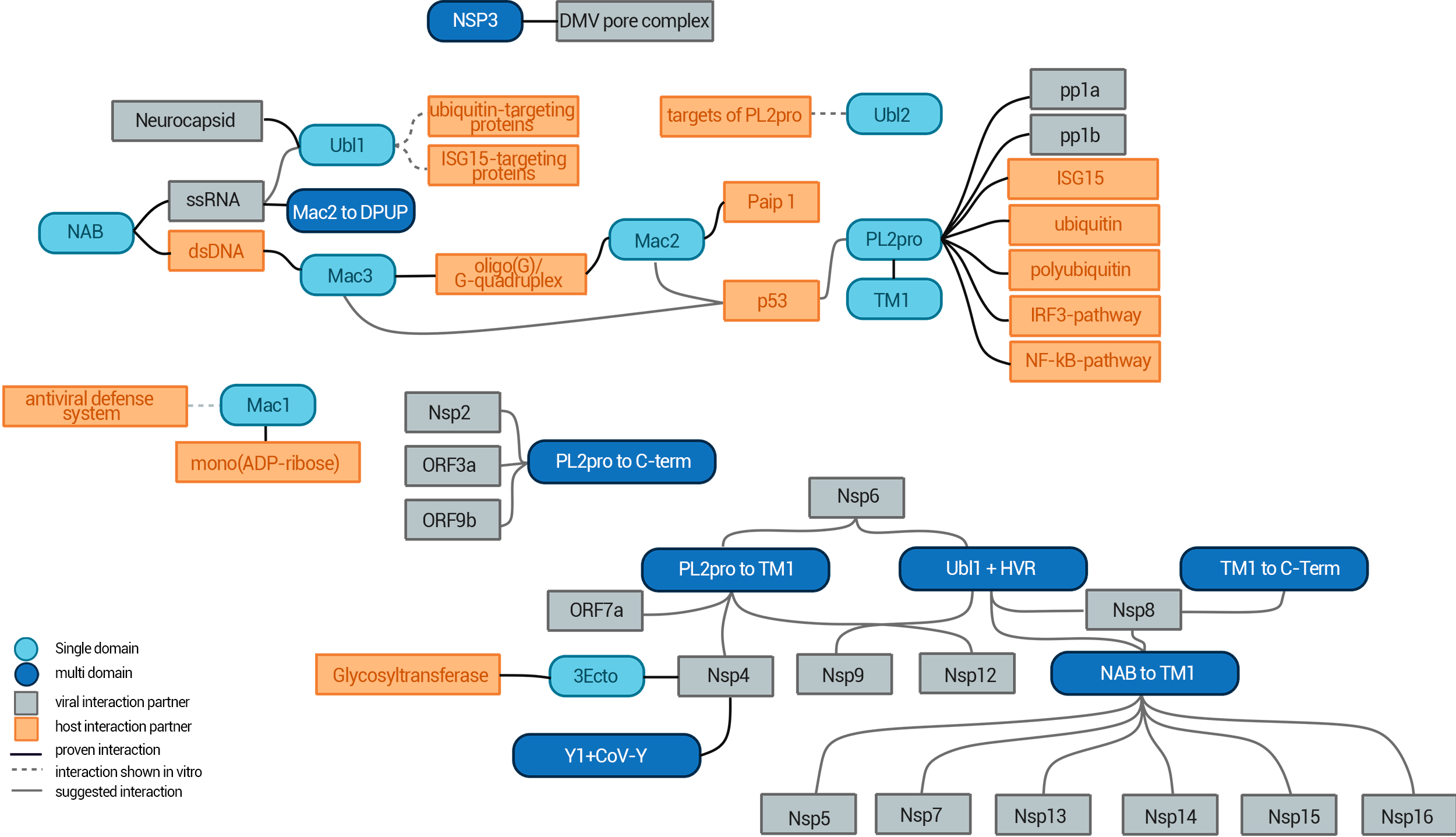
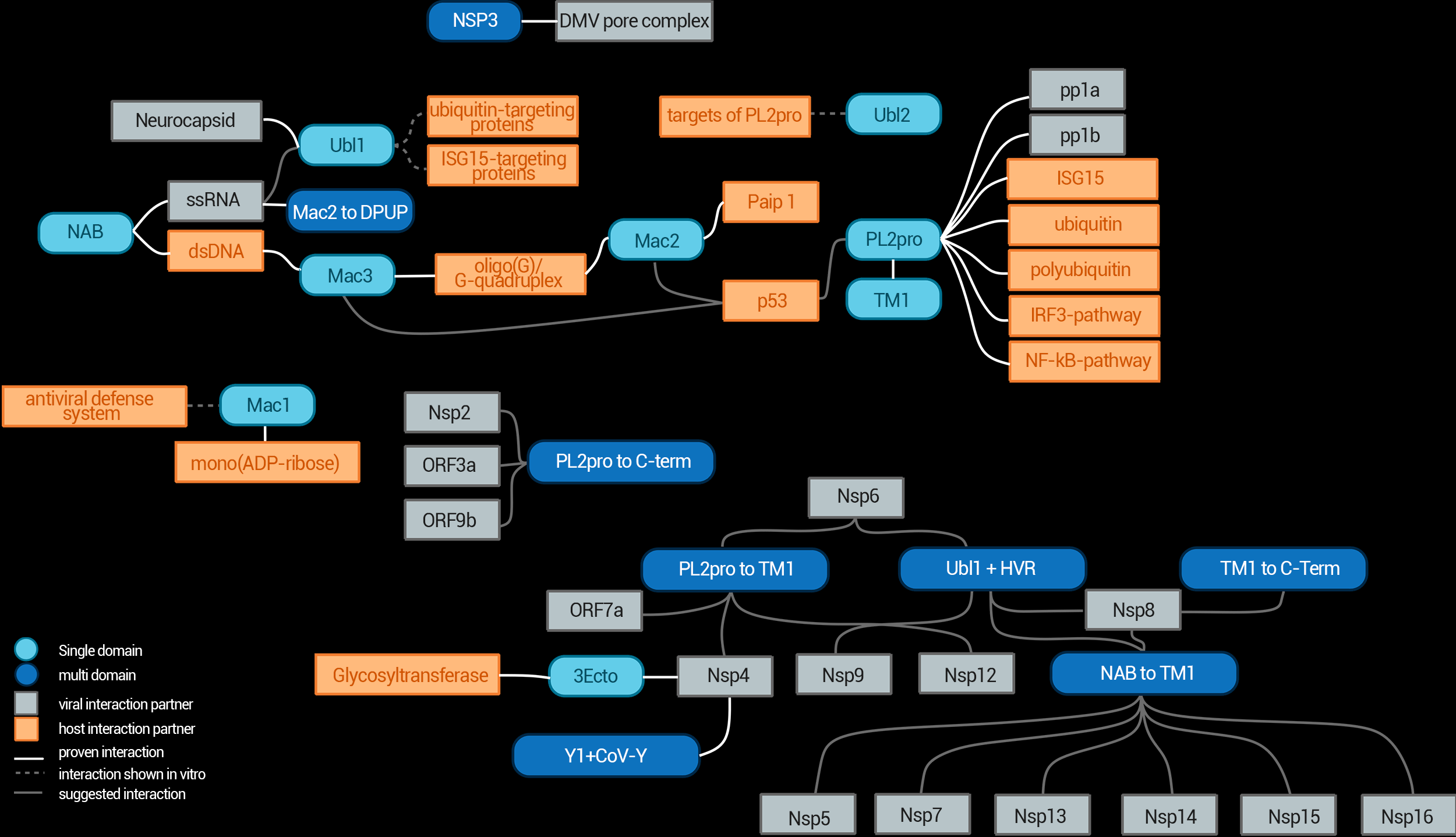
NSP3 Interaction Network
The interaction network of NSP3 and its individual domains.
Creator: Coronavirus Structural Task Force - Maximilian Edich
License: cc-by-sa
alternate image:
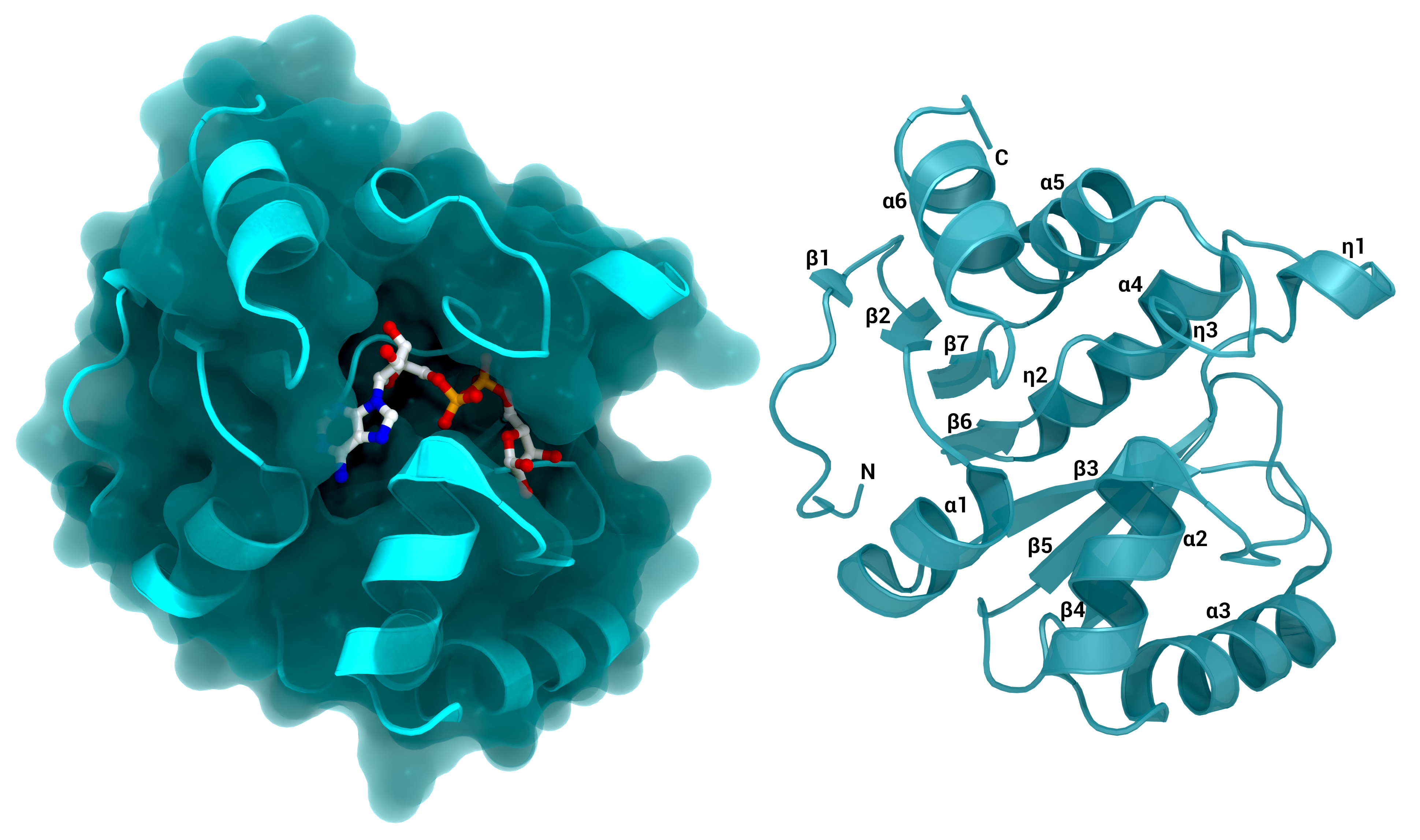
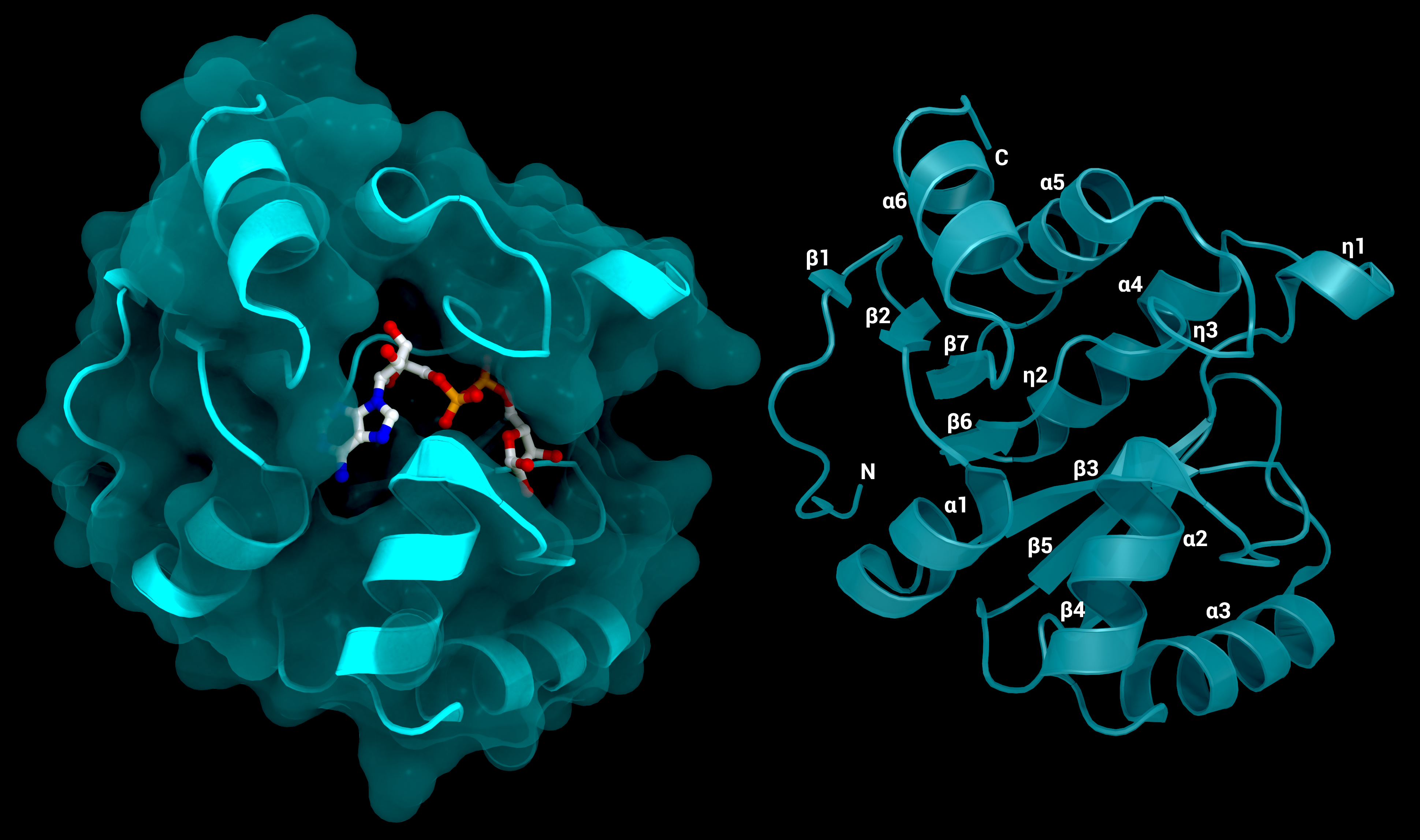
NSP3 Macrodomain 1
The SARS-CoV-2 macrodomain 1 of NSP3. Left: with surface and its substrate ADP-ribose, PDB ID: 7KQP. Right: labelled structure without substrate (PDB ID: 7KQO).
Creator: Coronavirus Structural Task Force - Protein Imager, Lea C. von Soosten
License: cc-by-sa
alternate image:
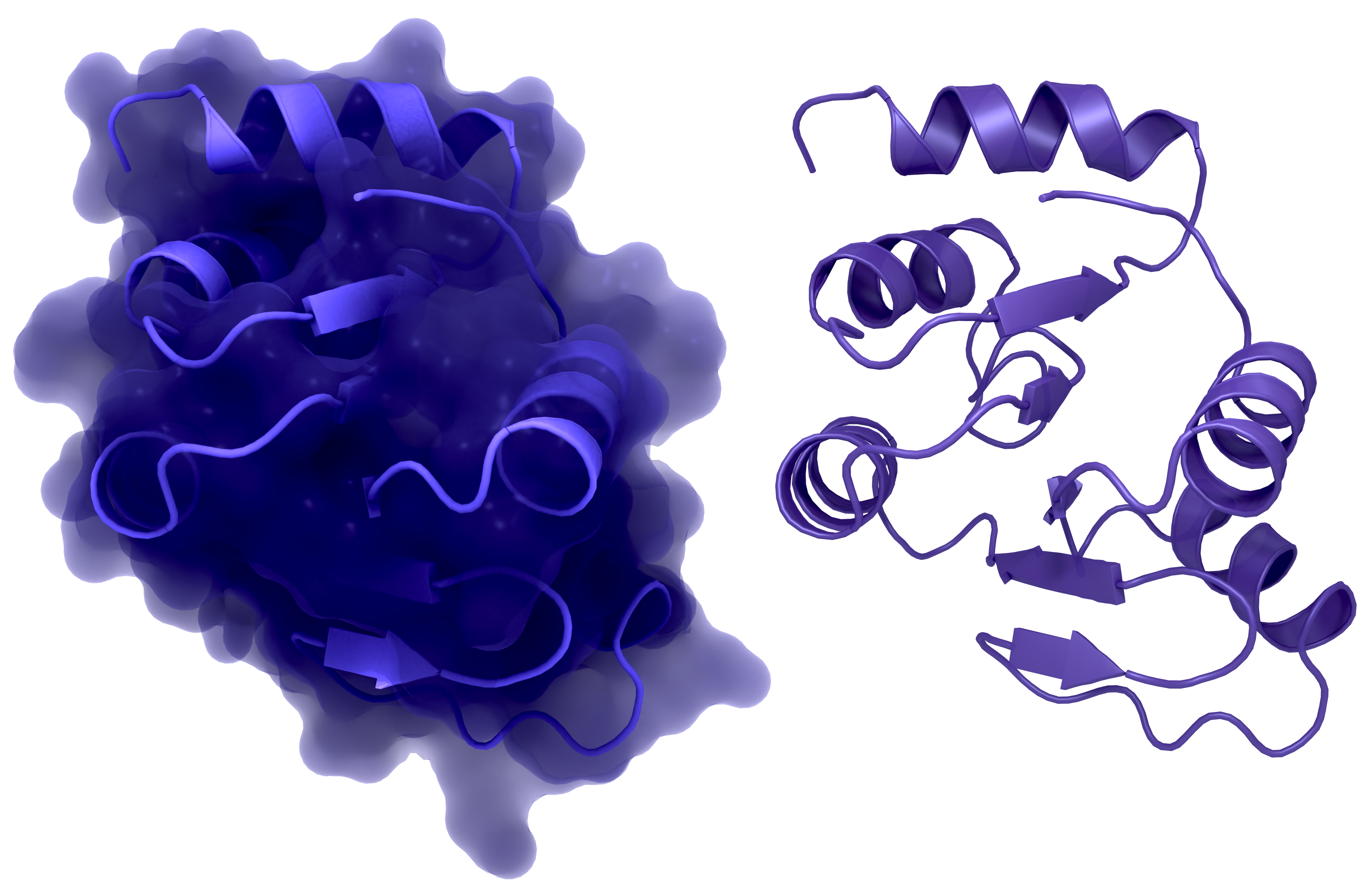
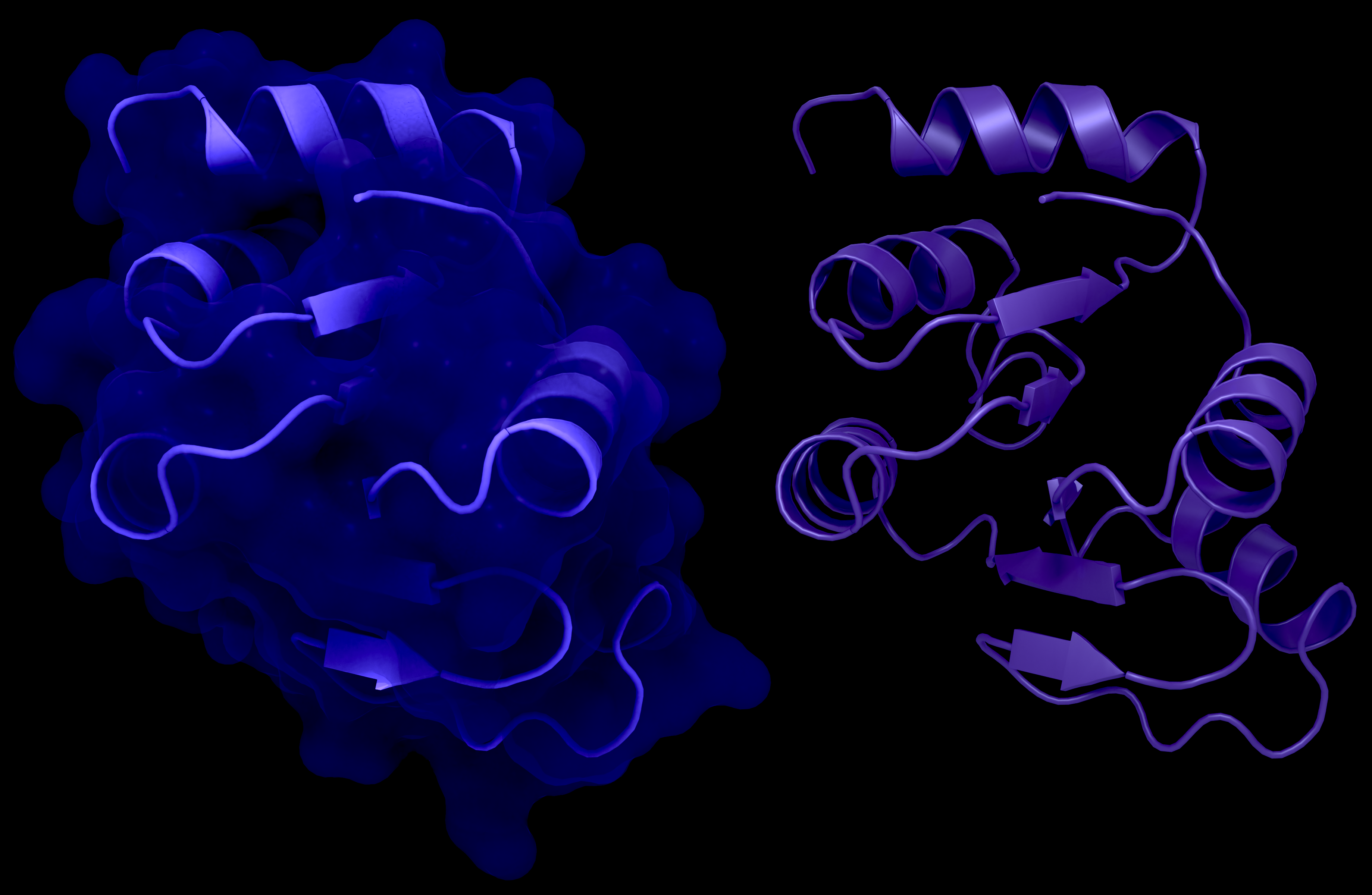
NSP3 Macrodomain 2
NSP3c or SUD of SARS-CoV-1: a: Structure of macrodomain 2 with (left) and without surface (right), PDB ID: 2W2G, amino acids 389-516 . b: Structure of macrodomain 3 with (left) and without surface (right), PDB ID: 2W2G, amino acids 527-652. c: Structure of the DPUP with (left) and without surface (right), PDB ID: 2KQW.
Creator: Coronavirus Structural Task Force - Protein Imager, Lea C. von Soosten
License: cc-by-sa
alternate image:
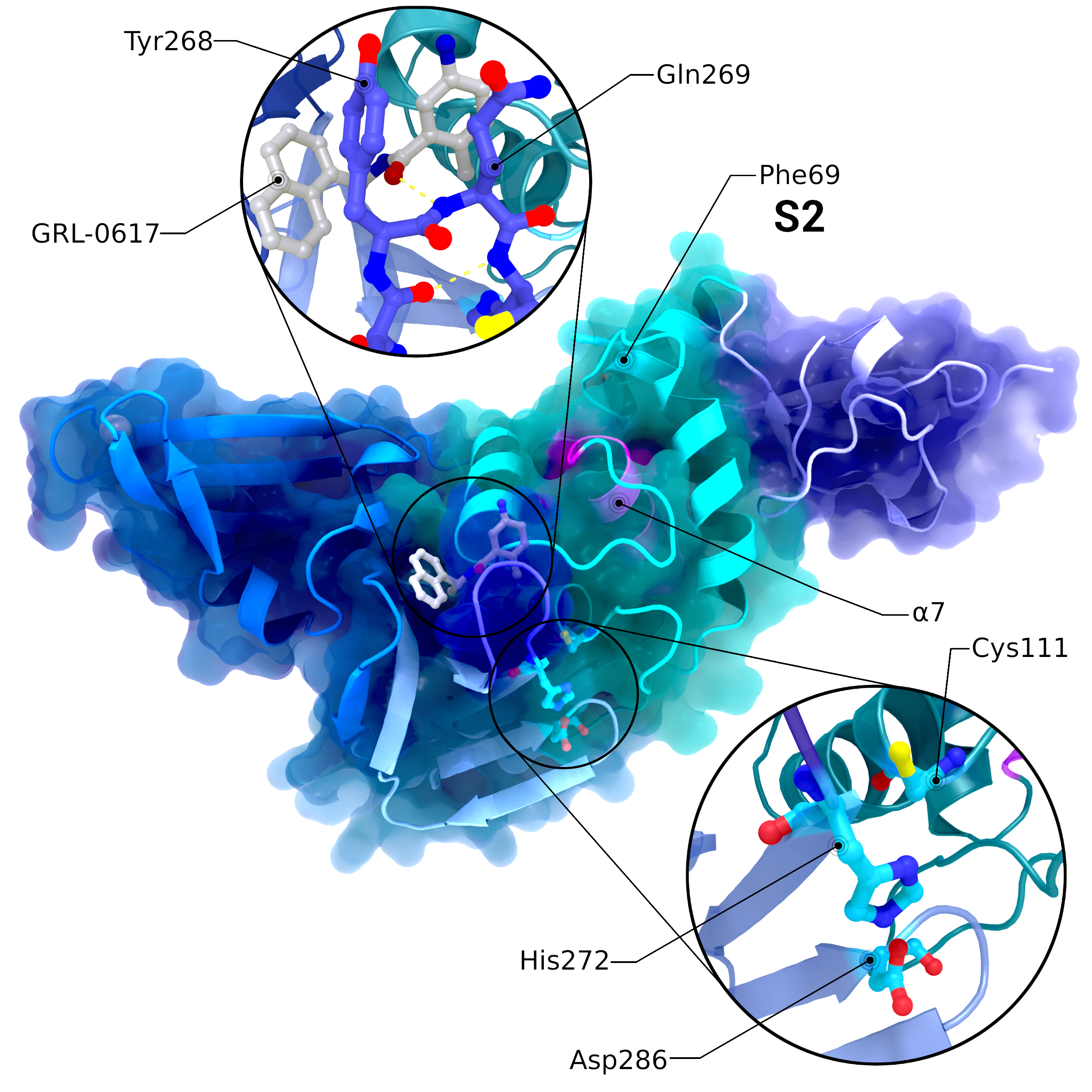
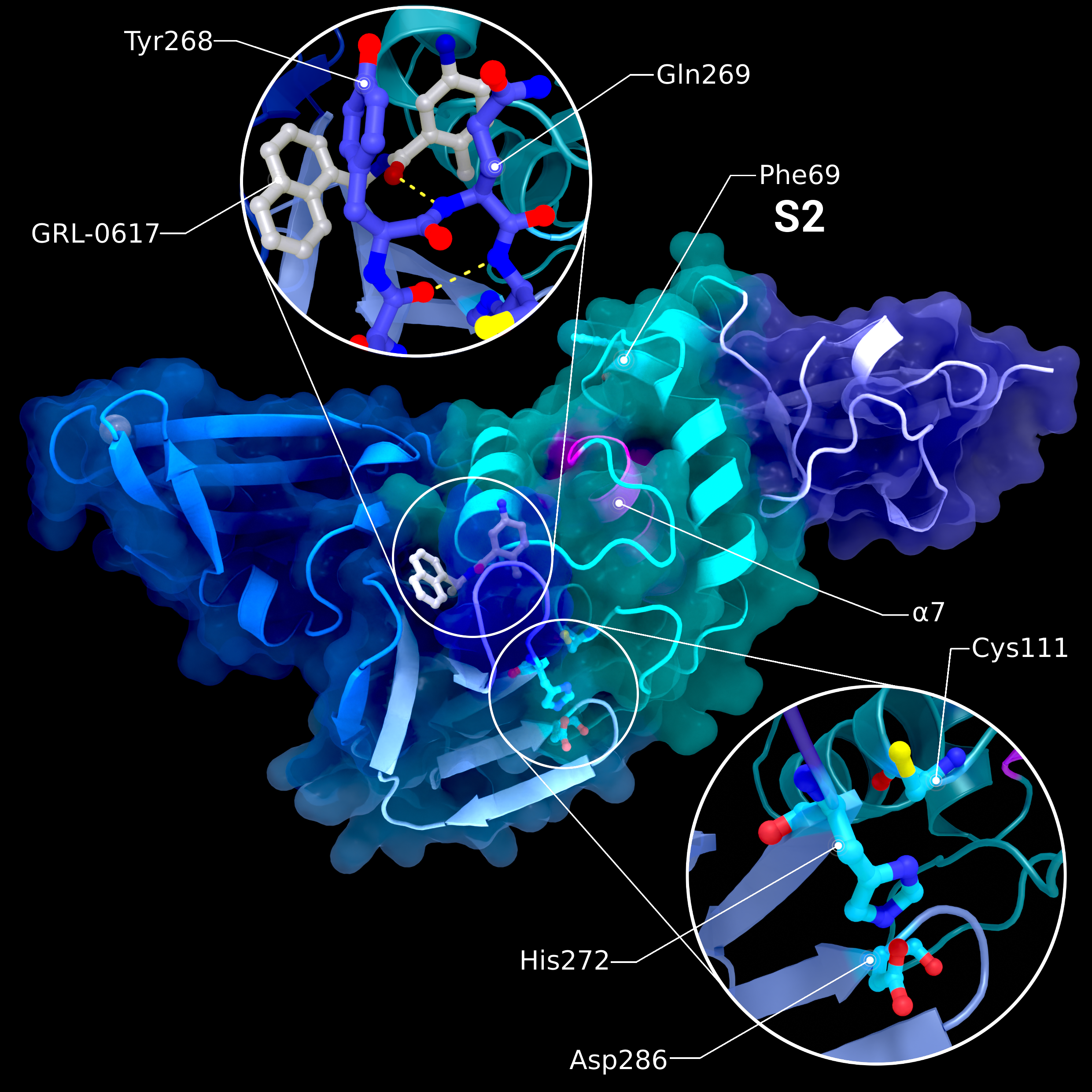
NSP3 PL2pro
Structure PDB ID 7JRN of PL2pro with attached Ubl2 domain (purple) and bound to inhibitor GRL-0617. In addition to Ubl2, PL2pro consists of the fingers- (dark blue, left), thumb- (cyan, right), and palm-subdomains (light blue, bottom). The catalytic active site is highlighted in the lower introspection, the blocking loop interacting with the inhibitor is highlighted in the upper introspection. S2 binding site is located at Phe69. Location of S1 varies depending on the substrate and is therefore not labelled. The pink helix α7 interacts with ISG15 binding at S1.
Creator: Coronavirus Structural Task Force - Protein Imager, Lea C. von Soosten
License: cc-by-sa
alternate image:
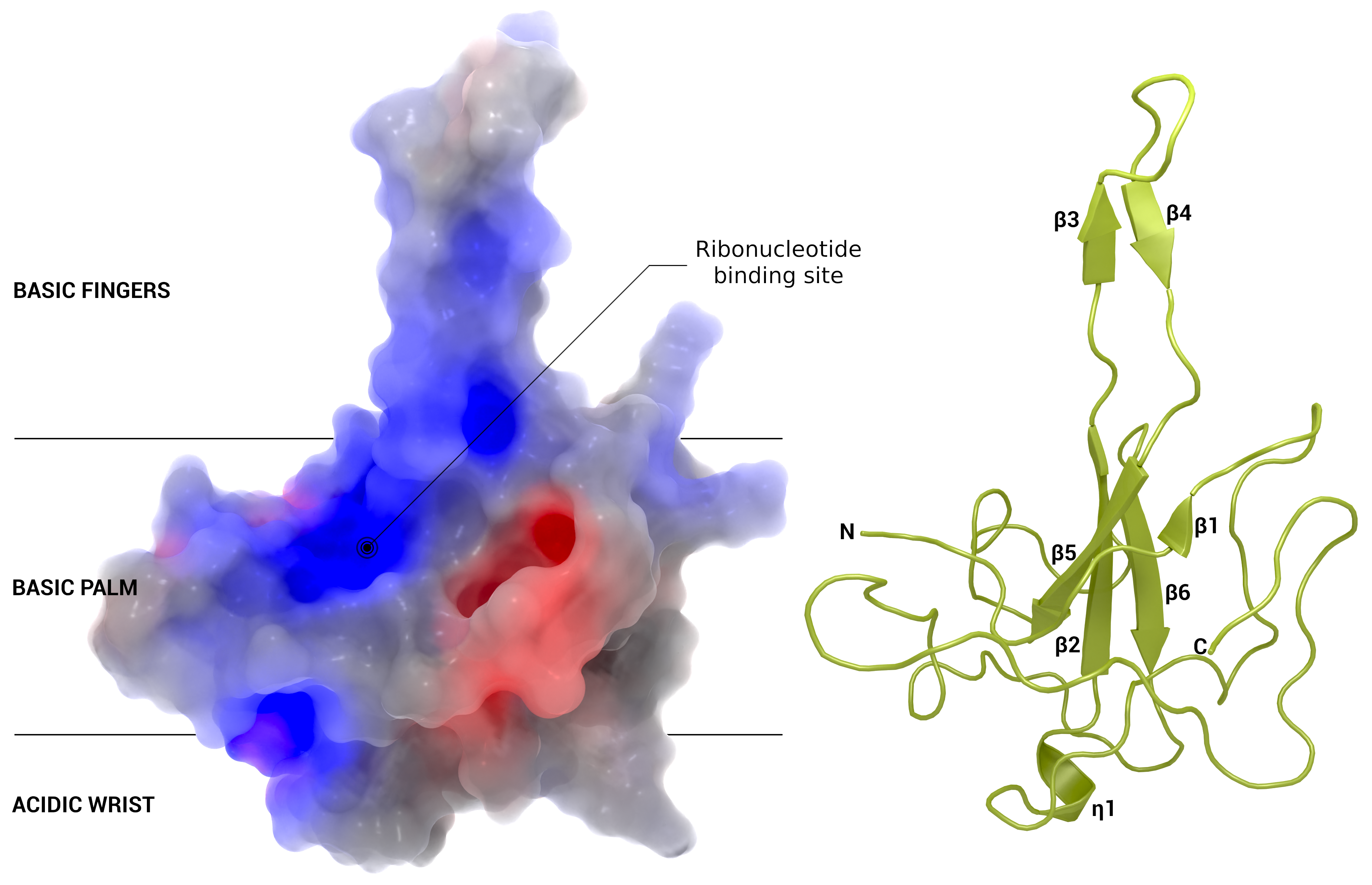
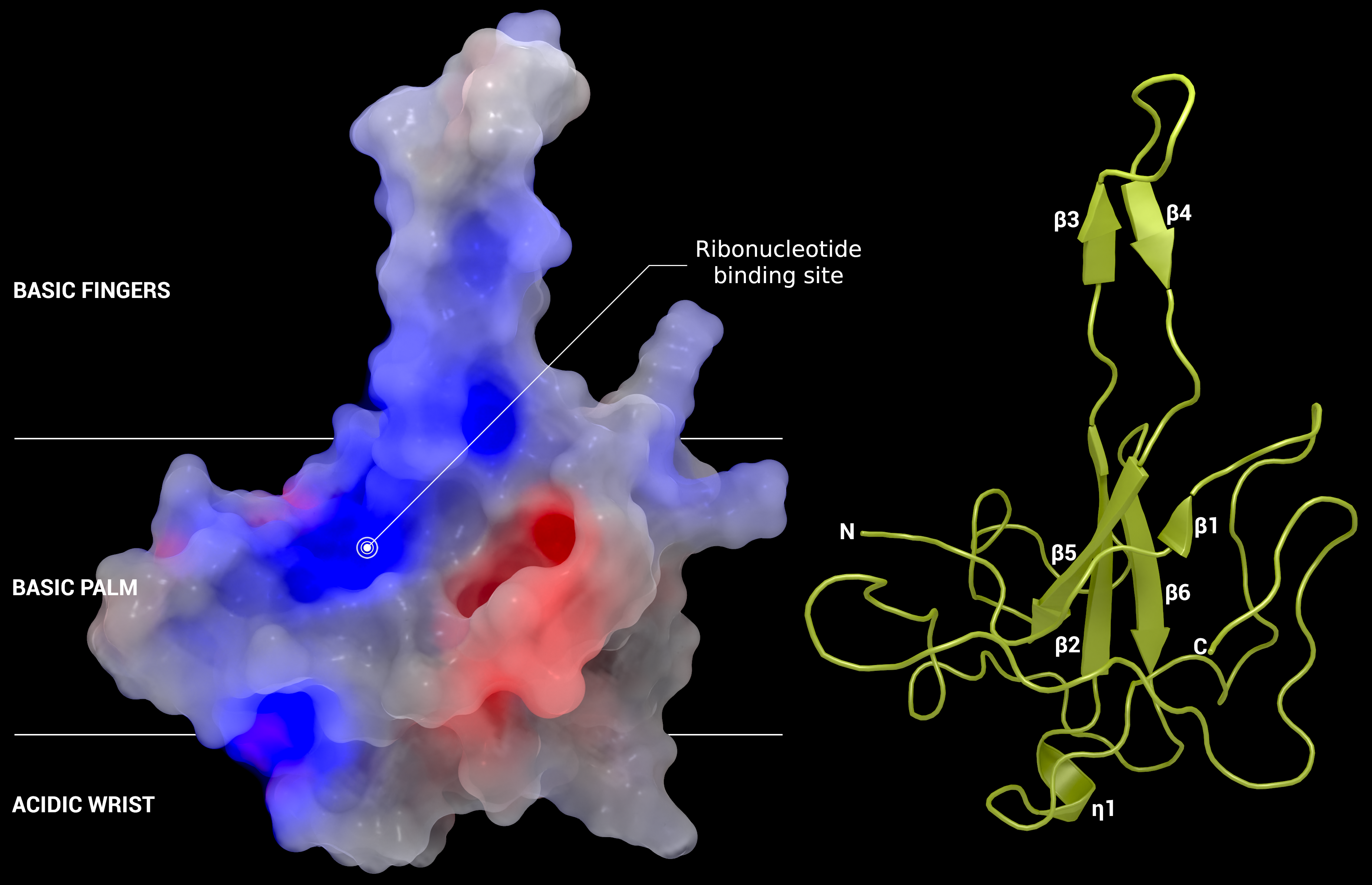
Nucleocapsid
Section of the nucleocapsid with possible binding site for active substances.
Creator: Coronavirus Structural Task Force - Protein Imager, Oliver Kippes
License: cc-by-sa
