“The coronavirus has led to a worldwide crisis for over a year. In a new study, nanoscientist Prof. Dr. Roland Wiesendanger illuminates the origins of the virus. His findings conclude there are a number of quality sources indicating a laboratory accident at the Wuhan Institute of Virology as the cause of the current pandemic.”
This is the beginning of an official press release from the University of Hamburg. Unfortunately, this study is not a study at all, but a rather confusing piece of internet research. And because we, the Coronavirus Structural Task Force, conduct research at the very same University of Hamburg, we would like to comment on the press release and this "study".

Here are the main arguments of the author, Roland Wiesendanger:
#1: Failure to identify the interim host proves that the disease is not of animal origin.
“In contrast to early coronavirus-based epidemics such as SARS and MERS, the scientific community has yet to identify the interim host that made the transmission of SARS-CoV-2 from bats to humans possible. Thus, there is no sound basis for a zoonotic theory as a possible explanation for the pandemic.” That an interim host of a zoonosis is not immediately identified is not unusual. The route of transmission of SARS-CoV was not elucidated until more than three years after the SARS pandemic [1]; for MERS, which was first described in 2012, it took two years [2] and to date some information is still missing [2–4]. Evidence indicates that interim hosts for SARS-CoV-2 may have been snakes, turtles, or pangolins [5–7]. Furthermore, transmission between humans and various animal hosts has been demonstrated several times (which, for example, unfortunately led to the culling of many mink)[8]. The lack of knowledge of an intermediate host does in no way disprove zoonosis as a cause.
#2: SARS-CoV-2 is so adapted to humans that it could not have arisen naturally
“The SARS-CoV-2 viruses are astonishingly effective at binding to human cell receptors and infecting human cells, thanks to its special cell receptor binding domains combined with a special (furin) cleavage site of the coronavirus spike protein. This is the first time a coronavirus has had both of these characteristics and indicates a nonnatural origin of the SARS-CoV-2 pathogen.”
The ability of the virus to bind to human cells with the spike protein is not evidence that this was artificially produced. Influenza, HIV, and Ebola are also all very good at binding to human cells [9] - the latter two have been shown to be animal-derived (zoonotic) pathogens. All of these viruses possess a furin cleavage site. Furins are enzymes found in all vertebrates that allow viruses to better target vertebrate cells. Thus, it is not unusual for SARS-CoV-2 to have evolved such a site through natural mutation and selection. In fact, it occurs naturally in many other coronaviruses as well [10].
The receptor binding domains S1 and S2 are highly variable because it is exactly domains that enable specific binding to host cells - the new mutations that are currently causing us so much trouble display variations right here. Mutations in this domain arise due to selection pressure in humans (or other hosts) and are also not indicative of a non-natural origin [11,12].
What also speaks against the virus being completely man-made, is that the sequence of the virus does not fit known methods for artificially producing genetic material [13,14]. Furthermore, designing a coronavirus would be considerably costlier than designing many other viruses because the genome is so large. This however, does not exclude the virus originating from a "gain-of-function" study.
Here it is perhaps worth noting that Professor Wiesendanger authored the study alone and is himself a non-specialist [15]. He has never published on Corona before, and therefore it is understandable that he is unfamiliar with both the details of genetic engineering and the technical terminology.
#3: Bats do not fly 2000km
“There were no bats for sale at the wet market in the center of Wuhan, which is the suspected hub of the outbreak. The Wuhan Institute of Virology, however, houses one of the largest collections of bat pathogens in the world, taken from distant caves in southern Chinese provinces. It is extremely unlikely that bats naturally made their way to Wuhan, from almost 2,000 km away, to then start a worldwide pandemic in the immediate vicinity of the Wuhan Institute of Virology.”
Considering that the thesis supported by WHO [16] is that there must have been an intermediate host, as is indeed also stated in the study and the press release (see 1.), the presence of bats near the first human vectors is not necessary. Many of the possible intermediate hosts were traded in the market in question. So far, there is no certainty that the pandemic originated there - research is still ongoing - but the absence of bats does not argue against zoonosis. The collection of bat viruses at the Center for Emerging Infectious Diseases in Wuhan does exist however. A researcher at this institute - Shi Zhengli - discovered that SARS originated from bats, and she conducted a systematic study of bat viruses from fecal samples in 2013, for example [17]. While the samples in question have been found in a cave 2000 kms from Wuhan, the bat in question (Rhinolophus affinis) does occur far and wide, including 250 km from Wuhan, in Hunan province. Bats have long been considered the largest source of different coronaviruses and thus the greatest risk for their transmission to humans [18].
#4: Research on viruses as bioweapons was conducted in Wuhan
“One research group at the Wuhan Institute of Virology had been researching the genetic manipulation of coronaviruses for many years with the goal of making these more infectious, more dangerous, and more fatal. This has been demonstrated by numerous publications.”
The publications cited in the study, for example this one [19], are indeed concerned with the recombination of bat coronaviruses with spikes that can bind to human cells. These were used to trace how the 2002/3 SARS pandemic originated - not to make the virus more dangerous. Such research has taken place elsewhere - with many strict safety measures and safeguards - for example in North Carolina [20].
#5: The virological institute in Wuhan was not secure.
“Safety measures were documented as being insufficient at the Wuhan Institute of Virology prior to the outbreak of the coronavirus pandemic.”
The laboratory in Wuhan is a Biosafety Level 4 laboratory [21], the highest level - there are only a handful of such laboratories in the world, of which two are in China. Such labs have strict access controls, you have to be able to hermetically seal them off, and they are under negative pressure to prevent pathogens from escaping; access is only through an airlock; all wastewater is chemically and thermally treated; a full protective suit has to be worn, and when leaving, the whole body has to be cleaned with soap. Of course, no laboratory is perfect, but safety is a paramount issue in such laboratories [21]. Nature has written a special report on the lab in Wuhan that illustrates this. In the text, Prof. Wiesendanger argues with a serious article from the Washington Post that points to episodes of malpractice in the laboratory in Wuhan, but whose sources remain undisclosed. Another source, a Youtube video, supposedly proves the improper disposal of laboratory equipment. However, while garbage can be clearly seen, typical laboratory waste such as pipette tips, consumables and gloves, is missing. Furthermore, the Chinese text of the video does not mention the waste in any way, it is about whether and how one could get into the building. A second source [22] shows a bat allegedly being sampled without protective equipment. Parts of the video, which is about a new coronavirus in pigs, show the non-BSL-4 area of the lab. The new coronavirus in pigs is said to come from the bat species shown. Nothing in this video suggests biosecurity problems in the lab.
6. Authorities covered up a laboratory accident in October
“There are numerous direct indications that the SARS-CoV-2 pathogen is of laboratory origin and point to a young researcher at the Wuhan Institute of Virology as being the first person to be infected. In addition, there are indications that the SARS-CoV-2 pathogen emerged from the Wuhan Institute of Virology into the city of Wuhan and beyond. There are also indications that the Chinese authorities conducted an examination of the institute in the first half of October 2019.”
The "indications of an official examination in the first half of October" are based on an analysis of mobile phone location data that an external firm is said to have produced for the Pentagon to show disruption of laboratory operations and road closures. However, the report provides no concrete evidence and was therefore deemed insufficient by the intelligence community. Especially since some of the allegations could be directly refuted [23]. Little can be found on the Internet about the young scientist, Yan Ling Huan, apart from a Twitter account, videos, and a rebuttal from the lab. The hypothesis that she was the first to be infected can therefore neither be proven nor disproven.
All in all, one cannot rule out the lab as the point of origin - research is being done on coronaviruses there - but the sources cited in this "study" are not evidence of that. Circulation of the virus before December cannot be ruled out either, but the publication does not present any robust evidence.
Additional notes
The main problem with this study is that it appears to have been written by the author alone and was not peer reviewed. This is particularly regrettable since the author emphasizes the importance of peer review on page 3. Apparently, discussion and especially media attention are very desirable, but peer review is not, which is why the "publication" was done on the Research Gate platform, where you can simply upload a PDF.

Other problems with the "study" are the poor readability and deficiencies in the evidence - this document is not only confusing, but also does not comply with good scientific practice. Sources include not only private communications and Twitter posts, but also content from the Alt-Right movement, such as an unrefereed study [24,25] from Steve Bannon's entourage, articles from the Epoch Times [26] or Summit News [22]. The document is also full of contradictions – for example, the first patient is said to have fallen ill on the first of October 2019, but elsewhere it says that the pandemic is due to a laboratory accident between October 6 and 11, 2019. The use of colored markers in the text also does not necessarily contribute to readability and is uncommon in publications. It is not clear why such a highly decorated and well-known scientist as Prof. Wiesendanger considered such writing publishable or even a study; and, unlike many of his colleagues, he does not do research on the coronavirus.
Conclusion
What comes across as an invitation to debate is a rather disorderly and angled piece of internet research that does not correspond with good scientific practice. Many people who are against China, or who are simply looking for someone to blame for Corona, feel vindicated and the University of Hamburg backs this up.
It is good and right that professors at German universities can publish and research whatever they want ("Forschungsfreiheit"). But the fact that this article is published in close consultation with the president [15] but without peer review and in the name of the university does not cast a good light on the University of Hamburg, which is, after all, our very own scientific home. Our statements here are about the shortcomings of this press release as well as the paper, and are not intended as criticism of Prof. Wiesendanger personally; we deeply regret this press release. As scientists, we should educate, critically question - and allow ourselves to be questioned. For the last twelve months, we have been informing colleagues and the general public about ongoing corona research, taken care not to accept funding from potential influencers (e.g. the pharmaceutical industry), and carefully reviewed every blog post, no matter how small. Every day we respond to inquiries from people who are confused, afraid of corona or of the measures being put in place. We teach, educate, we answer questions.
And that is what we will continue to do.
I would like to thank Dr. Florian Platzmann, Dr. Sam Horrell, Dr. Yunyun Gao, Pairoh Seeliger, Lea von Soosten, Katharina Hoffmann, Joshua Ezika and Sabrina Stäb for their help writing this article. Their expertise in lab safety, public health care, mandarin, molecular biology and their comprehensive internet / literature research made this opinion piece possible.
- [1]Li W, Wong S-K, Li F, et al. Animal Origins of the Severe Acute Respiratory Syndrome Coronavirus: Insight from ACE2-S-Protein Interactions. JVI 2006: 4211–4219. doi:10.1128/jvi.80.9.4211-4219.2006
- [2]Han H-J, Yu H, Yu X-J. Evidence for zoonotic origins of Middle East respiratory syndrome coronavirus. Journal of General Virology 2016: 274–280. doi:10.1099/jgv.0.000342
- [3]Middle East respiratory syndrome coronavirus (MERS-CoV). . Im Internet: https://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov); Stand: 19.02.2021
- [4]Chen F, Cao S, Xin J, et al. Ten years after SARS: where was the virus from? J Thorac Dis 2013; 5 Suppl 2: S163–S167. doi:10.3978/j.issn.2072-1439.2013.06.09
- [5]Ji W, Wang W, Zhao X, et al. Cross‐species transmission of the newly identified coronavirus 2019‐nCoV. J Med Virol 2020: 433–440. doi:10.1002/jmv.25682
- [6]Liu Z, Xiao X, Wei X, et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS‐CoV‐2. J Med Virol 2020: 595–601. doi:10.1002/jmv.25726
- [7]Zhao J, Cui W, Tian B. The Potential Intermediate Hosts for SARS-CoV-2. Front Microbiol 2020. doi:10.3389/fmicb.2020.580137
- [8]Mahdy MAA, Younis W, Ewaida Z. An Overview of SARS-CoV-2 and Animal Infection. Front Vet Sci 2020. doi:10.3389/fvets.2020.596391
- [9]Becker GL, Lu Y, Hardes K, et al. Highly Potent Inhibitors of Proprotein Convertase Furin as Potential Drugs for Treatment of Infectious Diseases. Journal of Biological Chemistry 2012: 21992–22003. doi:10.1074/jbc.m111.332643
- [10]Wu Y, Zhao S. Furin cleavage sites naturally occur in coronaviruses. Stem Cell Research 2021: 102115. doi:10.1016/j.scr.2020.102115
- [11]Lau S-Y, Wang P, Mok BW-Y, et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerging Microbes & Infections 2020: 837–842. doi:10.1080/22221751.2020.1756700
- [12]Liu Z, Zheng H, Lin H, et al. Identification of Common Deletions in the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus 2. J Virol 2020. doi:10.1128/jvi.00790-20
- [13]Almazán F, Sola I, Zuñiga S, et al. Coronavirus reverse genetic systems: Infectious clones and replicons. Virus Research 2014: 262–270. doi:10.1016/j.virusres.2014.05.026
- [14]Andersen KG, Rambaut A, Lipkin WI, et al. The proximal origin of SARS-CoV-2. Nat Med 2020: 450–452. doi:10.1038/s41591-020-0820-9
- [15]Ursprung des Coronavirus: Uni mit fragwürdiger Theorie. . Im Internet: https://www.zdf.de/nachrichten/politik/corona-labortheorie-universitaet-hamburg-100.html; Stand: 19.02.2021
- [16]WHO-convened Global Study of the Origins of SARS-CoV-2. . Im Internet: https://www.who.int/publications/m/item/who-convened-global-study-of-the-origins-of-sars-cov-2; Stand: 19.02.2021
- [17]Ge X-Y, Li J-L, Yang X-L, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013: 535–538. doi:10.1038/nature12711
- [18]Anthony SJ, Johnson CK, Greig DJ, et al. Global patterns in coronavirus diversity. Virus Evolution 2017. doi:10.1093/ve/vex012
- [19]Ren W, Qu X, Li W, et al. Difference in Receptor Usage between Severe Acute Respiratory Syndrome (SARS) Coronavirus and SARS-Like Coronavirus of Bat Origin. JVI 2007: 1899–1907. doi:10.1128/jvi.01085-07
- [20]Menachery VD, Yount BL Jr, Debbink K, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 2015: 1508–1513. doi:10.1038/nm.3985
- [21]Cyranoski D. Inside the Chinese lab poised to study world’s most dangerous pathogens. Nature 2017: 399–400. doi:10.1038/nature.2017.21487
- [22]Video From 2018 Shows Chinese Scientists Working On Coronavirus In Wuhan Lab. . Im Internet: https://summit.news/2020/04/30/video-from-2018-shows-chinese-scientists-working-on-coronavirus-in-wuhan-lab; Stand: 19.02.2021
- [23]Report says data suggests shutdown at China lab, but experts skeptical. NBC News. Im Internet: https://www.nbcnews.com/politics/national-security/report-says-cellphone-data-suggests-october-shutdown-wuhan-lab-experts-n1202716; Stand: 19.02.2021
- [24]Coronavirus: Wie Steve Bannon die Labor-Theorie verbreitet. . Im Internet: https://www.zdf.de/nachrichten/panorama/coronavirus-wuhan-labor-studie-yan-bannon-100.html; Stand: 19.02.2021
- [25]Yan L-M, Kang S, Guan J, et al. Unusual Features of the SARS-CoV-2 Genome Suggesting Sophisticated Laboratory Modification Rather Than Natural Evolution and Delineation of Its Probable Synthetic Route. 2020. doi:10.5281/ZENODO.4028830
- [26]Exclusive: China Had COVID-Like Patients Months Before Official Timeline. The Epoch Times (Singapore) 2020. Im Internet: https://epochtimes.today/exclusive-china-had-covid-like-patients-months-before-official-timeline/; Stand: 19.02.2021
Introduction
The coronavirus pandemic hit the entire world and caused millions of deaths. More than fifty companies race towards developing a vaccine to stop the disease (1). Vaccination presents a lasting solution to this unfavourable situation, reducing the burden of Coronavirus (2). The first vaccine to be approved for emergency use is an mRNA-based vaccine (3,4). How does it work? Here, I will shed some light on this, but before we go into details, we need to discuss a little about vaccines and how a foreign substance provokes an immune response in the body (immunogenicity).
Early impact of vaccines on humanity
Vaccines have existed since 1796 and have saved millions of lives over the years (5,6). Vaccines need to be given orally, intramuscularly or subcutaneously to, stimulate the body for an immune response and generate lasting immunity.
How the body fights diseases
The human body works very hard to remove any foreign substance (potential pathogens). When pathogens, such as bacteria or viruses enter the body, they attack and spread, causing disease. The immune system fights these infections recognizing a part of the pathogen using white blood cells which consist primarily of macrophages, B-lymphocytes and T-lymphocytes (7). Macrophages are antigen-presenting cells that engulf pathogens and digest them. Macrophages present parts of the pathogens to the T-lymphocytes and B-lymphocytes. When the B-lymphocytes are activated, they produce antibodies that target specific pathogens and destroy them. Also, when the T-lymphocytes are activated by the antigen-presenting cells, they seek out and destroy any infected cell in the body. However, it takes a couple of days for the body to generate the antibodies to fight an infection. After recovering from an infection, the immune system can remember how it fought the infection and can quickly in the future, prevent reinfection by the same or similar diseases. When the infection is gone, the body is left with the supply of memory cells called T-lymphocytes and B-lymphocytes that are responsible for this rapid intervention (7). Vaccines cause the same process but without a serious illness and often even without an infection. Individuals react to vaccines differently, some persons experience symptoms like swelling or redness at the injection site, low-grade fever, tiredness etc. These minor symptoms result as the body tries to build immunity against the disease, and it is your body`s healthy response. It would be no good and impracticable for the vaccine to cause the full-blown disease - instead, vaccines prime the immune system to jump into action should you ever encounter the real thing.
Traditional vaccines
Traditional vaccines contain either an attenuated (live or weakened) virus, an inactivated virus, or even a piece of a viral protein that can produce the immune answer – a so-called antigen that does not cause an actual disease but still stimulates the body to produce antibodies to fight a real infection (8). It tricks the immune system into thinking that an infection has occurred and the immune system responds by producing antibodies against the virus (9). mRNA-vaccines, however, do not contain dead or live pieces of the virus – they contain the genetic instructions necessary for our cells to make copies of the antigen itself. So, what is mRNA?
What is mRNA?
mRNA is how our body encodes information from the genome that is to be used as blueprints to make proteins: In every human cell, we find a nucleus that contains our genome in the form of DNA. This DNA is transcribed into mRNA, which then can leave the nucleus and enter the rest of the cell. DNA itself cannot leave the nucleus and this protects the cell’s genome from damage and manipulation. Once outside the nucleus, the mRNA is translated into proteins which then fulfil all kind of work tasks in our cells – they make up our hair, break down our food, transport oxygen around our body (10,11). They pretty much do everything that makes us go.
An mRNA vaccine exploits this process: When the mRNA vaccine enters our cells, it effectively skips the transcription step (DNA to RNA) and goes straight to translation (mRNA to proteins) to produce antigenic proteins (10,12).
How do the SARS-CoV-2 mRNA-based vaccines work?
The best antigen in the coronavirus SARS-CoV-2 – the bit which is recognized by the immune system – is the spike protein. The outside of the SARS-CoV-2 (Coronavirus) is carries these spike proteins, which the virus uses to enter human cells and cause an infection.
New mRNA vaccines contain messenger RNA which encodes the spike protein and as it enters the cell, the spike proteins are produced within the cell. The spike protein then migrate to the surface of the cell, where the immune system recognizes it as foreign and the body will produce antibodies to fight the infection. At this point, the process to produce immunity is the same as for any other vaccine. When infected again, the body can rapidly supply memory cells- T-lymphocytes and B-lymphocytes that remember how to attack a similar pathogen. So, when the real coronavirus SARS-CoV-2 (spike proteins and all) enters the body, the immune system remembers how it fought the formerly produced spike proteins and can easily fight off the infection.
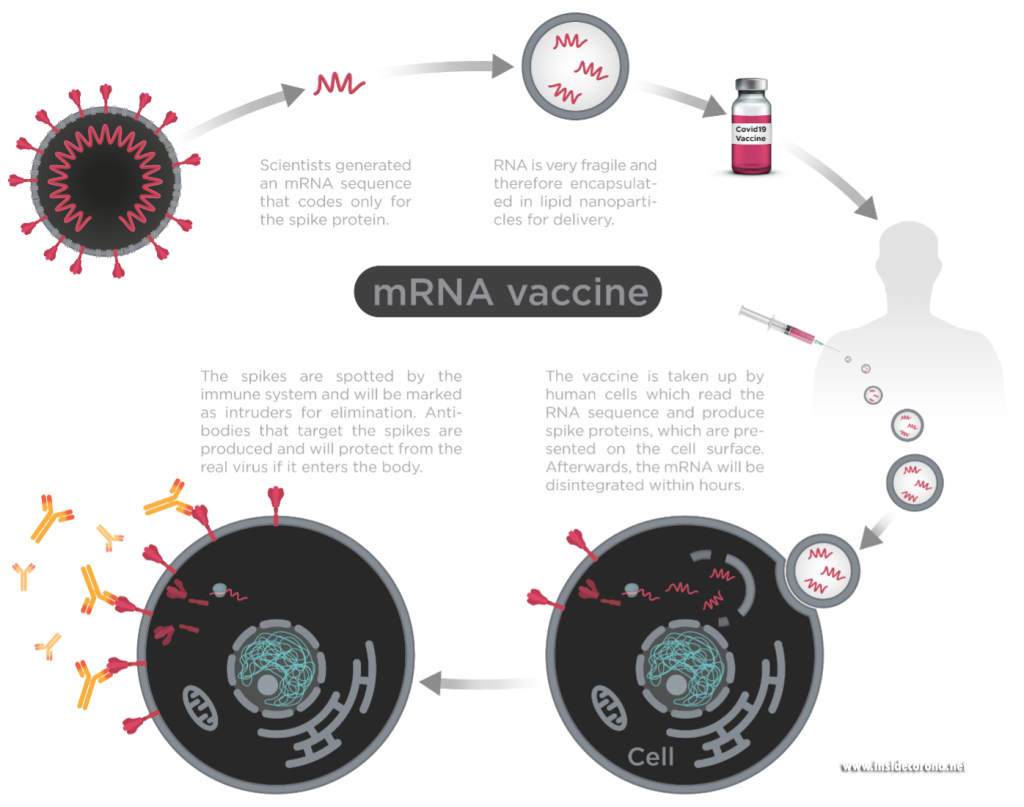
Figure1: Process of mRNA vaccines in the body.
Image by Thomas Splettstösser.
What is in the vaccine and why do we use it?
mRNA-based SARS-CoV-2 vaccines contain the mRNA strands encapsulated in lipid nanoparticles (think soap bubble) to protect it from hot temperatures as well as degrading enzymes (13,14). After being injected intramuscularly, the protection offered by the lipid nanoparticles helps the mRNA to remain active/potent until it enters the cells and releases mRNA.
There are some advantages to mRNA vaccines, and I think it can be expected that we will soon see more of them for other illnesses. The production of mRNA vaccine doses is much faster and cheaper than traditional vaccines since it does not require a long process of growing viral proteins in a cell or an egg which then needs to be deactivated or killed to produce the vaccine (15). Also, in mRNA vaccines there is never a complete virus, so contracting the disease is impossible, which can happen with live vaccines. Finally, should mutations occur, it is relatively straightforward to change the mRNA vaccine to contain this new mutation.
However, the storage temperature of an mRNA vaccine is a challenge: they need to be stored at a very low temperature to maintain its potency until it is ready to be used and this type of freezer is not available everywhere and makes transport difficult compared to a traditional vaccine (14,16).

Figure2: Sample storage freezer for mRNA vaccines.
Image by Yunyun Gao.
Can an mRNA-based vaccine change my DNA?
As the vaccine contains no so-called “reverse-transcriptase”, the spike mRNA cannot be converted into DNA. mRNA-based vaccines cannot even enter the cell’s nucleus. Hence, they are not able to change your DNA. They are just a messenger to produce the spike protein and the enzymes of the cell readily destroy them shortly afterwards. Also, the unfortunate cell itself is destroyed by the immune system once it displays the spike on the surface and the immune system has learned to recognize it.
Do I still need to wear masks?
Also, remember that after getting vaccinated, your body needs time to develop antibodies in sufficient quantities, so you can still contract Coronavirus even after being vaccinated. Hence, we need to maintain safety measures (social distancing, masking, handwashing etc) while allowing immunity to develop after being vaccinated. It is also still unclear what happens if an infected person is vaccinated, or in people who have no strong immune system (immunocompromised patients). “I have been vaccinated, can I now go, hugging and kissing on the street?” Sorry, you can’t. This is because you can still contract the virus even after being vaccinated: the body needs time to develop immunity against an infection. This is the main reason for jab-spacing as it enables the body to build an immune response.
Conclusion
Vaccines trigger the production of memory cells (T-lymphocytes and B-lymphocytes) to fight infections and can protect us from life-threatening diseases. The more people get vaccinated, the more likely we are to achieve herd-immunity, where even unvaccinated people are protected. A good example of herd-immunity is a burning bush. The fire keeps spreading but when it encounters a large space or a river, it stops spreading and the other side of the bush will not be affected. However, the space must be large enough to make this happen. Vaccinated individuals are like the large space or river, they help in stopping the spread of the disease to the unvaccinated hence, protecting the unvaccinated. Also, remember that for this kind of protection to occur, a sufficient number of individuals must have been vaccinated. There is no scientific basis to show that an mRNA-based vaccine can change your DNA. Developing an mRNA vaccine within a short period is a phenomenal advancement in science. As people are being vaccinated, every other protective measure is encouraged if we want to end this pandemic. We can only think of relaxing measures when the number of cases has considerably reduced. Stay safe and healthy, and together we are protected.
Acknowledgement
I would like to thank Dr Andrea Thorn, Dr Dale Tronrud, Dr Sam Horrell and Dr Yunyun Gao for their wonderful and helpful suggestions. Also, my thanks go to Dr Thomas Splettstoesser for making an image used for this post.
References
1. Regulatory Affairs Professionals Society. COVID-19 vaccine tracker [Internet]. [cited 2021 Jan 10]. Available from: https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker
2. Nature Communications. Vaccines work. Nat Commun [Internet]. 2018 Apr 24 [cited 2021 Jan 3];9. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5915378/
3. FDA. COVID-19 Vaccines. FDA [Internet]. 2021 Feb 18 [cited 2021 Feb 19]; Available from: https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines
4. Mueller B. U.K. Approves Pfizer Coronavirus Vaccine, a First in the West. The New York Times [Internet]. 2020 Dec 2 [cited 2021 Feb 19]; Available from: https://www.nytimes.com/2020/12/02/world/europe/pfizer-coronavirus-vaccine-approved-uk.html
5. World Health Organization. Vaccines and immunization [Internet]. [cited 2021 Jan 10]. Available from: https://www.who.int/health-topics/vaccines-and-immunization#tab=tab_1
6. Thèves C, Biagini P, Crubézy E. The rediscovery of smallpox. Clinical Microbiology and Infection. 2014 Mar;20(3):210–8.
7. Vitetta ES, Berton MT, Burger C, Kepron M, Lee WT, Yin XM. Memory B and T Cells. Annual Review of Immunology. 1991;9(1):193–217.
8. Centers for Disease Control and Prevention. Basics of Vaccines | CDC [Internet]. 2019 [cited 2021 Jan 10]. Available from: https://www.cdc.gov/vaccines/vpd/vpd-vac-basics.html
9. Sell S. How vaccines work: immune effector mechanisms and designer vaccines. Expert Rev Vaccines. 2019 Oct;18(10):993–1015.
10. Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines — a new era in vaccinology. Nature Reviews Drug Discovery. 2018 Apr 1;17(4):261–79.
11. Zhang C, Maruggi G, Shan H, Li J. Advances in mRNA Vaccines for Infectious Diseases. Front Immunol [Internet]. 2019 Mar 27 [cited 2021 Jan 11];10. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6446947/
12. Jackson NAC, Kester KE, Casimiro D, Gurunathan S, DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. npj Vaccines. 2020 Feb 4;5(1):11.
13. Martin C, Lowery D. mRNA vaccines: intellectual property landscape. Nat Rev Drug Discov. 2020 Sep;19(9):578–578.
14. Baden LR, Sahly HME, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. New England Journal of Medicine [Internet]. 2020 Dec 30 [cited 2021 Jan 10]; Available from: https://www.nejm.org/doi/10.1056/NEJMoa2035389
15. Sandbrink JB, Shattock RJ. RNA Vaccines: A Suitable Platform for Tackling Emerging Pandemics? Front Immunol. 2020;11:608460.
16. Pan American Health Organization WHO. COVID-19 Vaccine Explainer: COMIRNATY®, COVID-19 mRNA vaccine - PAHO/WHO | Pan American Health Organization [Internet]. [cited 2021 Feb 5]. Available from: https://www.paho.org/en/documents/covid-19-vaccine-explainer-comirnatyr-covid-19-mrna-vaccine
Overview
During the Corona-dominated year 2020 scientists all over the world united and gathered as much information as possible to understand the exact mechanism behind the lifecycle of SARS-CoV-2.
The main question was: how can we stop the virus from invading the human cell and causing COVID-19? A focus in the quest to answer this question, was the SARS-CoV-2 entry mechanism. The group of Janet Iwasa contributes to this ongoing research process by providing a high-quality video animation of the SARS-CoV-2 entry into the human host cell. This current version of the entry animation has already been shown on PBS News (08.12.20) and we aim to improve it with your help in 2021 (see below)!
The Entry Animation
Click this Link to see the Entry Animation on YouTube.
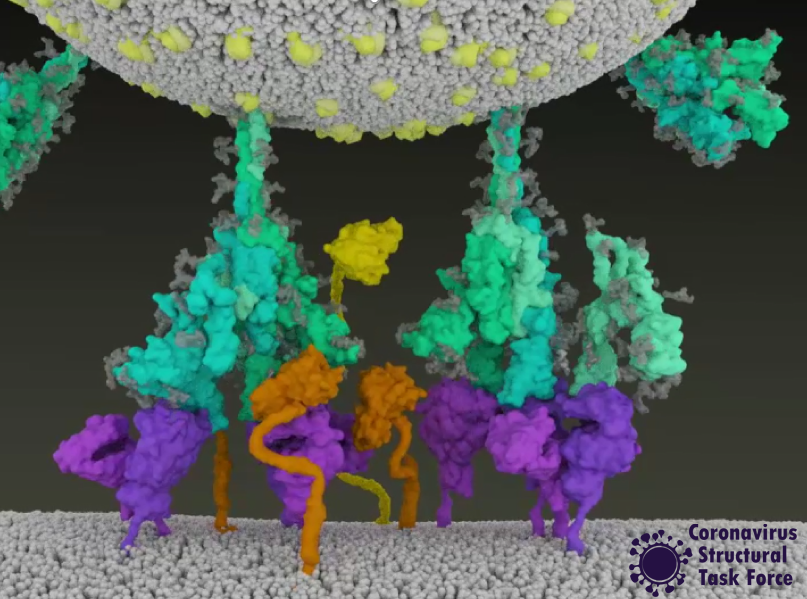
This entry animation is a collection of current knowledge about the SARS-CoV-2 entry mechanism. What we know at this point is that the mechanism starts with the viral approach. An individual can be infected with SARS-CoV-2 after inhaling airborne viral particles. These viruses can then travel into the airways, where they may encounter host cells of the respiratory epithelium in the trachea and lungs.
As you can read in a previous blogpost, the Spikes (teal) are Corona’s key to invade the host cell and thus of great interest in terms of vaccination and therapeutic approaches against COVID-19. The Spike protein recognizes a specific receptor on the human host cell surface, called ACE2 (purple). Usually, the Spikes are very dynamic and able to undergo opening, closing and bending movements. But after binding to ACE2, the protein is locked into its open position. Another protein on the cell surface, called TMPRSS2 (orange), can then come along and cut the Spike protein in a specific location. These segments of the Spike protein fall away, exposing a portion of the Spike protein which was previously hidden.
The Spike protein is then able to undergo a series of dramatic conformational changes. During the first stage, the Spike protein inserts itself into the membrane of the cell. In the second stage, segments of the Spike protein zipper back on itself, forcing the membrane of the cell and the viral membrane to fuse. After fusion, the viral RNA is deposited into the host cell, where it will direct the cell to produce more virions. This process is known as post-fusion.
The Annotation Tool
Click this Link to use the Annotation Tool.
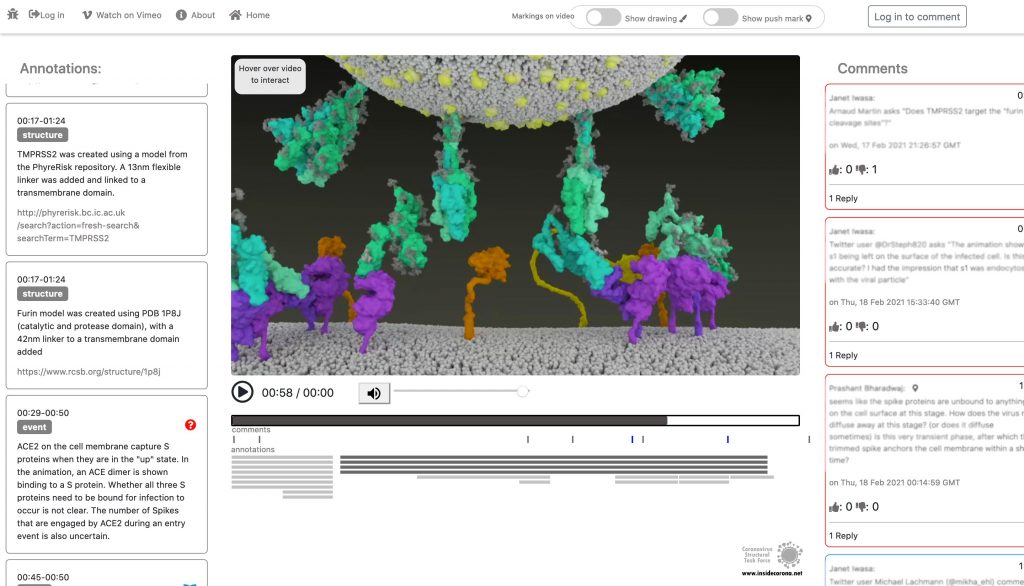
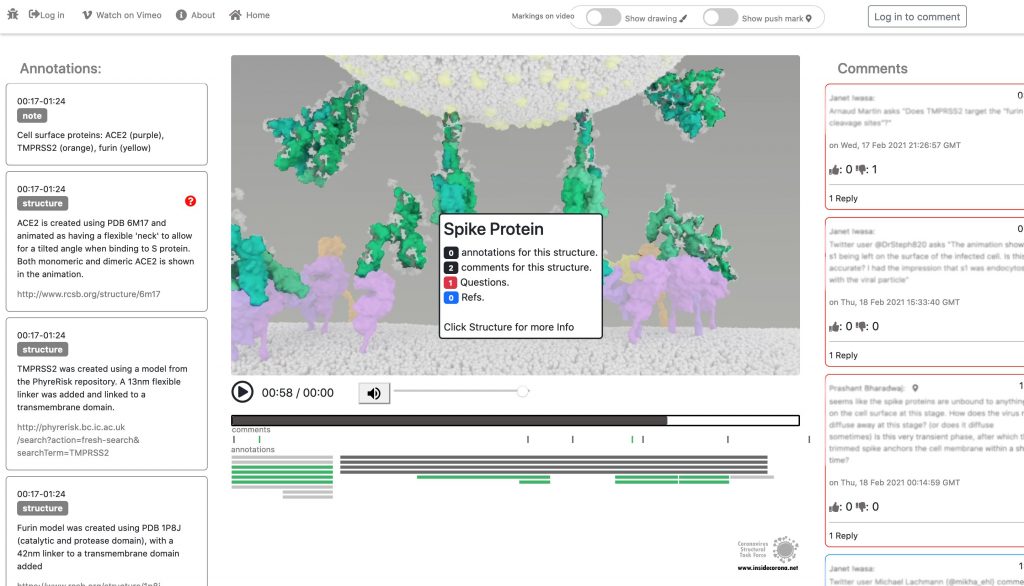
In January, this will be supplemented with a tool so that the knowledge about the SARS-CoV-2 entry mechanism can be discussed interactively by scientists all over the world. This online platform will serve as a basis for scientific discussion by providing an annotation tool. Scientific users can set a pin at any point of the video and comment their suggestions, criticism or questions about the mechanism and the structure depictions (see Fig. 1 for a prototype). Based on these annotations, the Iwasa Group will improve the animation of the entry process to provide an up-to-date detailed representation of this key process. The resulting entry animation is not only addressed to scientists, but it is also used for public outreach and education.
Even though the entry mechanism is not entirely understood yet, it could already be depicted in the fantastic animation of the Iwasa Group. There are still a lot of details and additional information to be found out about this process. From January on, the annotation tool therefore will provide the opportunity to discuss this mechanism publicly.
Thanks to the Iwasa Group for this Christmas present!
Merry Christmas!
Introduction
It is known as VUI‑202012/01 or B.1.1.7 – the new mutation of the coronavirus Sars-CoV-2. It may be responsible for a sharply increased number of infections in the southeast of England (1), however, the scientific results leading to very strict lockdown measurements in the south of the UK, and travel restrictions across Europe are few and far between. Here, we have compiled what is known up until now.
On mutations
Mutations are normal in the evolution of life – and of viruses. If two similar viruses have infected the same cell, their genomes can become mixed-up, one of the reasons why animal influenza strains are considered so dangerous. This is also called recombination. Mutations can be caused by chemicals, radiation (including UV light) and errors during genome copying. A typical SARS-CoV-2 virus accumulates two amino acid changes per month in its genome — a rate of change about half that of influenza (2). This is because SARS-CoV-2 can repair RNA to some extent. But even so, this natural process led to thousands of mutations since the beginning of the pandemic. If they affected the virus life cycle negatively, that strain may have likely died out - if they did not make a difference or enhanced its chances of survival, it may have persisted.
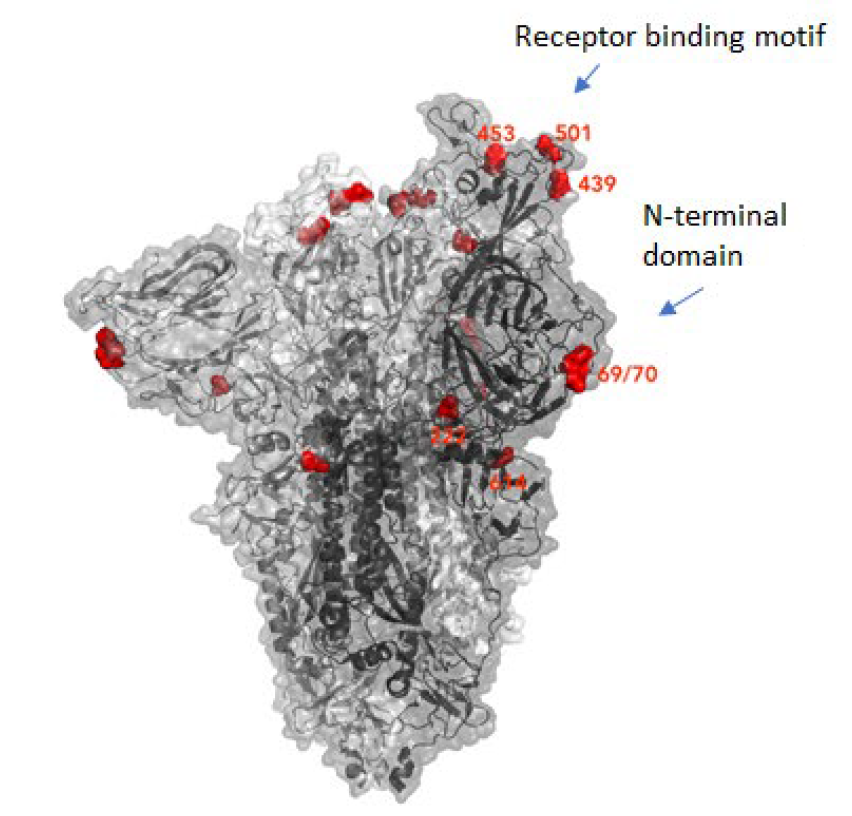
Many mutations that are observed occur in the spike protein, which both serves to recognize potential host cells but is also what is being recognized by antibodies (i.e., the immune system).
Changes here can be crucial for the survival of the virus (“evolutionary pressure”) as they could significantly alter its affinity to the human receptor ACE2, which the virus uses as gateway to our cells.
What vaccines do
Most, if not all, potential COVID-19 vaccines expose our body to some part of the spike protein, which can be made by the body itself (mRNA vaccines) or carried by a harmless virus instead of SARS-CoV-2 (vector). Our body then produces antibodies which specifically recognize the spike and persist for several months. If we are exposed afterwards to the real virus, the body can recognize it immediately – and the risk of infection is much lower as the immune system swings into action immediately. Earlier this year, the spike mutation D614G (amino acid residue number 614 changing from aspartic acid (D) to glycine (G)) caused quite a stir in the media, and became the predominant form of SARS-CoV-2 (2, 3). However, if and in how far this was caused by natural selection is still debated (3). Another example which triggered an increased media coverage was the mutation Spike Y453F, which originated from infected minks in Denmark (4) and led to a culling of millions of animals. In any case, if we would be vaccinated with a spike protein form that would be different from the one in a virus we encounter later, there is a small chance that the vaccine may be rendered ineffective. This chance is, however, small for SARS-CoV-2, in any case much smaller than for HIV, which famously evaded any attempt to develop a vaccine.

What do we know?
There was a steep rise in infections in the UK recently, as in most other European countries.
A new mutation of the virus has emerged and seems to replace the old version of SARS-CoV-2 (5). Thousands of patients have been found to carry this variant.
This new variant has more mutations at once than expected. These mutations have not observed in this combination before.
The variant has been reported in the UK, the Netherlands, Denmark, Australia and Belgium so far.
What is striking to me as scientist about these findings is one thing in particular: How could the British government find that thousands of people were having the new SARS-CoV-2 variant, instead of the old, if the illness does not look any different? Sequencing samples from each and every patient would be technically very challenging, if not impossible. How could they know? The answer is:
Serendipity
The main PCR test employed in the United Kingdom is Thermo Fisher's TaqPathCOVID-19. This test identifies RNA on three different genome locations: In ORF1ab, nucleotide and spike. Now, it stopped working for the spike portion of the test, while the other two RNAs were still found to be present, which likely prompted scientists to sequence some of the samples in question. And indeed, the new mutant has a deletion of histidine-69 and valine-70, called 69-70del. This permitted easy differentiation of patients with the old SARS-CoV-2 (3 hits) and the new (2 hits) and is the reason why we know so much about the epidemiology of this variant!* It has also to be said that this test is not used as often in other countries, such as Germany, and this could well be the reason why we do not know if and how widespread it is here. In addition, other countries sequence much smaller proportions of virus isolates than the UK, so ongoing circulation of this variant outside of the UK cannot be excluded.
The details of the mutation
The new variant of SARS-CoV-2 VUI-202012/01 has 14 amino acid changes and three deletions affecting the genes for ORF1ab, spike and ORF8. One of these mutations (N501Y) occurs in the receptor binding domain and could lead to an increased binding affinity to the human ACE2. The 69-70 deletion has likely an immunological role and is the reason this mutant was detected so widely, as this RNA location is used for PCR tests. Another interesting mutation is the P681H, which is next to a furin cleavage site that has a biological significance in membrane fusion. These mutations could be responsible for the increased transmissibility. The effects of the other mutations aren’t fully investigated yet. Here is a list of the mutations which have been observed in the VUI‑202012/01 or B.1.1.7 variant:
| T1001I in gene ORF1ab | |
| A1708D in gene ORF1ab | |
| I2230T in gene ORF1ab | |
| SGF 3675-3677 deletion in gene ORF1ab | |
| A1708D in gene ORF1ab | |
| HV 69-70 deletion in spike | The 69-70 deletion on the spike protein is a re-occurring mutation that has shown to often co-occur with other amino acid changes in the RBD (6, 7). (1) Evasion to the human immune response and in association with other receptor binding domain changes (1) (2) Immunological role (8) (3) Leads to diagnostic failures which permit detection (see above, "Serendipity") (4) Associated with immune escape in immunocompromised patients (9(8)) Furthermore, the 69-70 deletion arose in multiple unrelated lineages and is associated with the evasion of the immune response (9). It is being hypothesized that this mutation undergoes a strong positive selection when exposed to convalescent plasma therapy in an immunocompromised human host (7). |
| Y144 deletion in spike | Deletion in the spike N-terminal domain (9) |
| N501Y in spike | One of six key contact residues in the spike receptor binding domains, this mutation leads to an increasing binding affinity to human and murine ACE2 (1). |
| A570D in spike | Mutation located at the spike receptor binding domain (10) |
| P681H in spike | The P681H mutation is located directly next to the furin cleavage site. It is one of the four residues which are insertions when compared to closely related coronaviruses, creating a furin cleavage site in the spike protein between the spike S1 and S2 domains. This prompts the entry of the virus into respiratory epithelial cells as well as the transmission in animal models (1) The S1/S2 furin cleavage site of SARS-CoV-2 is not found in closely related coronaviruses and has been shown to promote entry into respiratory epithelial cells and transmission in animal models (9) |
| T716I in spike | Mutation in in the S2 domain |
| S982A in spike | Mutation in in the S2 domain (10) |
| D1118H in spike | Mutation in in the S2 domain (8) |
| Q27 stop in ORF8 | The Q27stop mutation in the ORF8 leads to the truncation of the ORF8, and as it only consists of 121 amino acids, the consequence might be a loss of function. These and the other mutations could be responsible for the increased transmissibility of the B.1.1.7 variant. In any case, this mutation truncates the ORF8 protein at residue 27 or renders it inactive which allows further downstream mutations to accrue. (1) |
| R52I in ORF8 | |
| Y73C in ORF8 | |
| D3L in nucleocapsid | |
| S235F in nucleocapsid |
Why were there so many mutations at once?
This could be a result of prolonged or chronical SARS-CoV-2 infections as study of these infections reveal unusually large numbers of nucleotide changes and deletion mutations and often high ratios of non-synonymous changes. In addition to this, convalescent plasma treatment can cause intra-patient virus genetic diversity (11).
What does the new mutation mean in terms of impact and epidemiology?
There was an increase in cases with the new strain in total and in
proportion to the old (1). What does that mean for us?
This is what the internet says:
The COVID-19 genomics UK consortium (COG) reports about a “priority set of SARS-CoV-2 Spike mutations that are of particular interest based on potential epidemiological significance in the UK and/or biological evidence based on the literature or unpublished work.” (9)
The New and Emerging Respiratory Virus Threats Advisory Group of the British government (NERVTAG) discussed the new variant on Friday and concluded that its growth rate is higher by 67-75% and that this is likely due to a selective advantage. “In summary, NERVTAG has moderate confidence that VUI-202012/01 demonstrates a substantial increase in transmissibility compared to other variants.” (12) This is very likely the source of Boris Johnson’s claim to this strain being “70% more infectious”.
The English government writes that PHE (Public Health England) „is working with partners to investigate and plans to share its findings over the next 2 weeks. There is currently no evidence to suggest that the variant has any impact on disease severity, antibody response or vaccine efficacy. High numbers of cases of the variant virus have been observed in some areas where there is also a high incidence of COVID-19. It is not yet known whether the variant is responsible for these increased numbers of cases.” (13)
Conclusion
From this, we conclude that the British government, and we, do not know yet. It has not been conclusively shown that the new variant is more infectious (likely), has an easier time to evade the host immune system or if the vaccine will be less effective against it (very unlikely). The epidemologic model which predicts a higher tranmissability has still to be published, the science is still in the making. Tests of vaccines against the new variant are ongoing and will take a few weeks. There is yet little evidence that this new variant poses a significantly bigger threat than others - or to the contrary.
Acknowledgements
While I am listed as author of this article, it could not have been written without the help and research by Pairoh Seeliger, Lea von Soosten, Luise Kandler, Erik Nebelung and Oliver Kippes who all helped in this.
I would also thank Nicolai Wilk from Thermo Fisher Scientific who quickly responded to my questions about their test.
The title picture shows mutation cards from the game Pandemic Expansion: On the Brink by Z-Man Games.
- *The 69-70del mutation is predominantly observed in B.1.1 (including B.1.1.7), B.1.258, and the cluster 5 variant lineages of SARS-CoV-2.
References
- 1.A. Rambaut, Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. virological.org (2020), (available at https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563).
- 2.E. Callaway, The coronavirus is mutating — does it matter? Nature, 174–177 (2020).
- 3.L. Zhang, C. B. Jackson, H. Mou, A. Ojha, H. Peng, B. D. Quinlan, E. S. Rangarajan, A. Pan, A. Vanderheiden, M. S. Suthar, W. Li, T. Izard, C. Rader, M. Farzan, H. Choe, SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun (2020), doi:10.1038/s41467-020-19808-4.
- 4.ECDC, Detection of new SARS-CoV-2 variants related to mink. www.ecdc.europa.eu (2020), (available at https://www.ecdc.europa.eu/sites/default/files/documents/RRA-SARS-CoV-2-in-mink-12-nov-2020.pdf).
- 5.ONS UK , Percentage of COVID-19 cases that are positive for ORF1ab and N genes. www.ons.gov.uk (2020), (available at https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/adhocs/12690percentageofcovid19casesthatarepositivefororf1abandngenes).
- 6.R. M. Dawood, M. A. El-Meguid, G. M. Salum, K. El-Wakeel, M. Shemis, M. K. El Awady, Bioinformatics prediction of B and T cell epitopes within the spike and nucleocapsid proteins of SARS-CoV2. Journal of Infection and Public Health (2020), doi:10.1016/j.jiph.2020.12.006.
- 7.S. A. Kemp, D. A. Collier, R. Datir, S. Gayed, A. Jahun, M. Hosmillo, I. A. Ferreira, C. Rees-Spear, P. Mlcochova, I. U. Lumb, D. Roberts, A. Chandra, N. Temperton, K. Sharrocks, E. Blane, J. A. Briggs, K. G. Smith, J. R. Bradley, C. Smith, R. Goldstein, I. G. Goodfellow, A. Smielewska, J. P. Skittrall, T. Gouliouris, E. Gkrania-Klotsas, C. J. Illingworth, L. E. McCoy, R. K. Gupta, Neutralising antibodies drive Spike mediated SARS-CoV-2 evasion (2020), , doi:10.1101/2020.12.05.20241927.
- 8.K. Kupferschmidt, Mutant coronavirus in the United Kingdom sets off alarms, but its importance remains unclear. Science (2020), doi:10.1126/science.abg2626.
- 9.COG, COG-UK update on SARS-CoV-2 Spike mutations of special interest Report 1. https://www.cogconsortium.uk (2020), (available at https://www.cogconsortium.uk/wp-content/uploads/2020/12/Report-1_COG-UK_19-December-2020_SARS-CoV-2-Mutations.pdf).
- 10.S. Kemp, W. Harvey, R. Datir, D. Collier, I. Ferreira, A. Carabelii, D. L. Robertson, R. K. Gupta, Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion ΔH69/V70 (2020), , doi:10.1101/2020.12.14.422555.
- 11.ECDC, Threat Assessment Brief: Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. www.ecdc.europa.eu (2020), (available at https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-rapid-increase-sars-cov-2-variant-united-kingdom).
- 12.NERVTAG, NERVTAG meeting on SARS-CoV-2 variant under investigation VUI-202012/01. https://khub.net (2020), (available at https://khub.net/documents/135939561/338928724/SARS-CoV-2+variant+under+investigation%2C+meeting+minutes.pdf/962e866b-161f-2fd5-1030-32b6ab467896?t=1608470511452).
- 13.PHE, PHE investigating a novel variant of COVID-19 . www.gov.uk (2020), (available at https://www.gov.uk/government/news/phe-investigating-a-novel-variant-of-covid-19).
The SARS-CoV-2 virus has a huge negative impact on our lives, a strong contrast to the incredible small size of the virus. This elusiveness poses major challenges to our understanding and ability to fight it. In times of fake news and politicians who are abusing the pandemic for their own agenda by claiming that the virus is a hoax, there is a valid question to ask: ‘How can we be sure that the virus exists if they are even too small to be seen under the microscope?’
This question describes a scientific problem that can be traced back to the 19th century, when the true nature of viruses was a mystery even to microbiologists and doctors.
Here, we will go on a hunt for the proof of virus’s virulent existence. We will discuss the two main indications scientists look out for in matters like this, and how the scientific community solved this mystery during the past two centuries:
Indication 1: Evidence by Causality
On the first step of our hunt let us have a look at the subject: Viruses! One infamous virus is the one that is responsible for rabies. Rabies has 100% lethality after the first symptoms occur. These symptoms include fever, intense pain and a digestive tract that goes haywire. Later symptoms include hyperactivity as well as hydrophobia. This last symptom leads to the ill person developing an aversion to swallow their own saliva and feeling uncomfortable around any type of water. A terrible disease that made doctors and scientists desperately searching for the cause. But even long after bacteria had been found and identified as the origin of certain diseases (culprits for diseases are also called pathogens), no such bacteria that could be held responsible for causing rabies were ever found.
Thus the scientists of the time were confronted with a question: What exactly was responsible for diseases like rabies? Was the pathogen comparable to bacteria, or just so very alien in nature that it would not compare to anything known until then? Was it even physical in nature?

One of the persons who came one step closer to the answer of these questions was the French microbiologist Luis Pasteur, who lived in the 19th century. Previous successes with anthrax and fowl cholera established him as an icon in medicine and microbiology. But his meticulous search for the pathogen responsible for rabies was fated to be unsuccesful from day one, since none of the responsible pathogens could ever have been seen with the microscopy techniques of the time.
Given the disease’s severe nature and lethality, Pasteur tried to develop a vaccine against rabies, despite the fact that there was no hint on a pathogen’s existence except the symptoms it caused. Drawing on his experience with bacterial pathogens he eventually succeeded to develop an effective vaccine. He circumvented the need to isolate the pathogen by using infected nerve tissue to pass the disease to different mammal species like rabbits and dogs, reducing its virulence with every cycle. Just a few years later after succesfully testing it on dogs - in 1885 - the vaccine safed the life of the 9-year-old French boy Joseph Meister, making him the first person ever to be immune to rabies.

So, even when the true nature of the rabies pathogen remained a mystery, Pasteur was still able to prevent it from infecting people (and dogs), just by carefully observing the matter at hand, and learning from the way bacteria work. The fact that this technique worked implied that this pathogen was interacting with our bodies in a similar way to bacteria, proving that it must be physical in nature and hence we must be able to find it.
The development of the Chamberland filter, by Charles Chamberland, one of Pasteur’s assistants at the time, narrowed the physical properties of the pathogen down further. The filter basically consists of a porcelain tube with an opening on one side through which a liquid can be filtered, provided sufficient pressure.
This filter was shown to separate observable microorganisms from the liquid, including pathogenic bacteria. In 1892, Russian biologist Dimitry Iwanowski, who was investigating the mosaic disease of certain plants, filtered juice from infected leaves with the Chamberland filter and documented that the extracted juice was still infectious to healthy tobacco plants. He also observed that, unlike disease-inducing bacteria which would lose their ability to survive after a short time without nutrients, the filtrate remained infectious for many months after the filtration process. This not only added further evidence to the true nature of viruses but incidentally foreshadowed one of viruses' most annoying traits: The ability to survive without any kind of metabolism, akin to plants, animals or bacteria.

This experiment was repeated with many other diseases in the following years, including rabies. Dutch microbiologist Martinus Beijerinck first introduced the word “virus” for the filtered infectious liquid in 1898 (From Latin virus: "poison, slime, venom"). And although scientist were still uncertain if viruses were liquid or composed of particles, these experiments demonstrated that the responsible pathogens must be soluble in water and small enough to pass the Chamberland filter. This is a prime example of how scientists can demonstrate the existence of something from its effects on its environment under controlled conditions, which may even permit educated guesses about its nature.
But as they say: “Seeing is Believing”, (not "Educated Guessing is Believing") so let’s proceed our hunt for specifically visible proof with:
Indication 2: Evidence by Visualization
In the years after the discovery of the infectious liquids called “viruses” microscopy techniques improved more and more, to the point where clumps of multiple virus particles could be shown. Some scientists supposedly saw these clumped viruses without knowing it to be viruses; but the resolution and magnification abilities of light microscopes were nowhere near high enough to determine any kind of structure. There is a physical limit to the magnification a microscope can achieve. To discuss this limit, we must digress a bit and take a closer look on how waves work, and how light and matter interact:
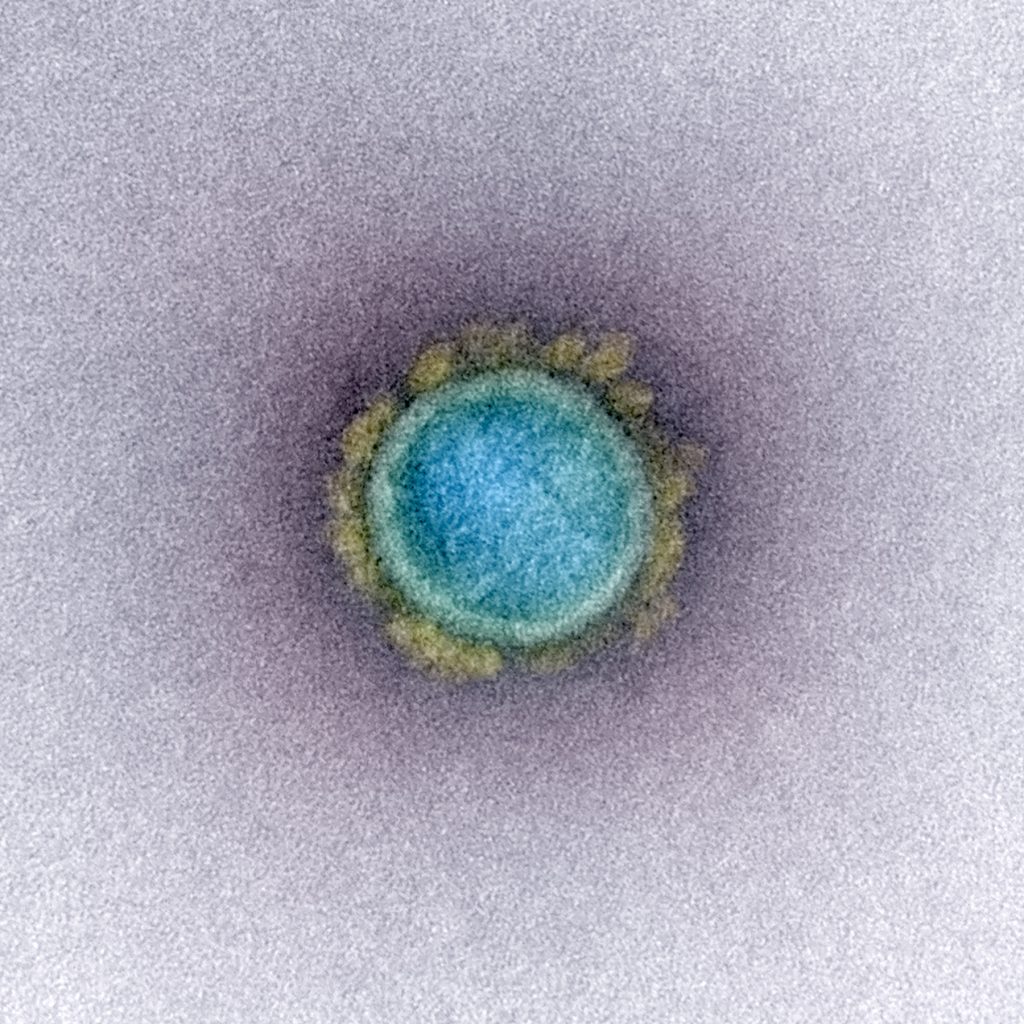
Light can be seen as a wave. Reflection, diffraction and refraction alongside things like absorption and emission are the main interaction mechanisms between this wave and matter and they allow us to see. However, all these mechanisms depend on some level on the compared size between the waves legth and the object. Smaller objects would just be “ignored” by the wave. Since waves of any kind share some of their basic principles, a comparison with water waves can come in handy to visualize this property: Imagine boulders disrupting the waves on a beach. One could deduce the boulder's existence and make assumptions on their size by the way the waves move past these boulders and arrive on the beach. Now imagine a 20 m high super wave, which would just sweep over them and not show any sign of their existence. Similarly, electromagnetic waves ignore obstacles that are much smaller than the wavelength.* This is the reason why viruses are being “ignored” by the light waves used in light microscopy. To be precise: The spectrum of visible light (which is most commonly used in light microscopy) covers wavelengths from roughly 700 nm down to ca. 400 nm. Considering the just discussed dependence of light-matter interaction on size and the way a microscope guides its light, adding the application of some math, we find the highest theoretical resolution in light microscopy to be calculated around 250 nm. Even today, however, a great expense of resources and effort must be spent to come close to this level of resolution. Most viruses are much smaller. The SARS-CoV-2 virus measures in at roughly 120 nm diameter, less than half the theoretical limit!
Now, since light microscopy cannot let us see viruses, what other way might there be to make them visible to us, on our hunt for the visible proof of viruses existence?
Enter the Electron Microscope:

This revolutionary device was first built by German scientists Max Knoll and Ernst Ruska in 1931. This was shortly after French physicists Lois de Broglie’s hypothesis that particles can also behave like waves went ‘viral’ in 1923. The electron microscope takes this concept and circumvents the electromagnetic wavelength problem by waving electromagnetic waves goodbye and using the wave behaviour of accelerated electrons instead. Here, the lenses which focus the light were replaced with magnetic lenses in order to focus electrons (which are charged negatively). These coil arrays produce magnetic fields that bend the pathway of the electrons. After having passed the sample, and another magnetic lens, the electrons are detected by a fluorescent screen, which translates the signals of the incoming electrons into signals we can see. Since one could theoretically accelerate the electrons to velocities very close to the speed of light - thereby shortening their wavelength more and more - the theoretical resolution limit for electron microscopes is basically unlimited.
However, relativistic effects, exponentially growing energy costs for acceleration close to the speed of light, and technical aspects of magnetic lenses limit the practical resolution limit of modern electron microscopes to some still mind blowing 0.1 - 0.2 nm.
The electron microscope opened doors to deeper and smaller levels of the microcosmos for the scientific community, and as fate would have it, the brother of Ernst Ruska, Helmut Ruska, happened to be a physician and biologist. Very soon, he realized his brother’s inventions potential for microbiology, and together with their colleague Bodo von Borries they published the first microscopic picture of a virus in 1938.
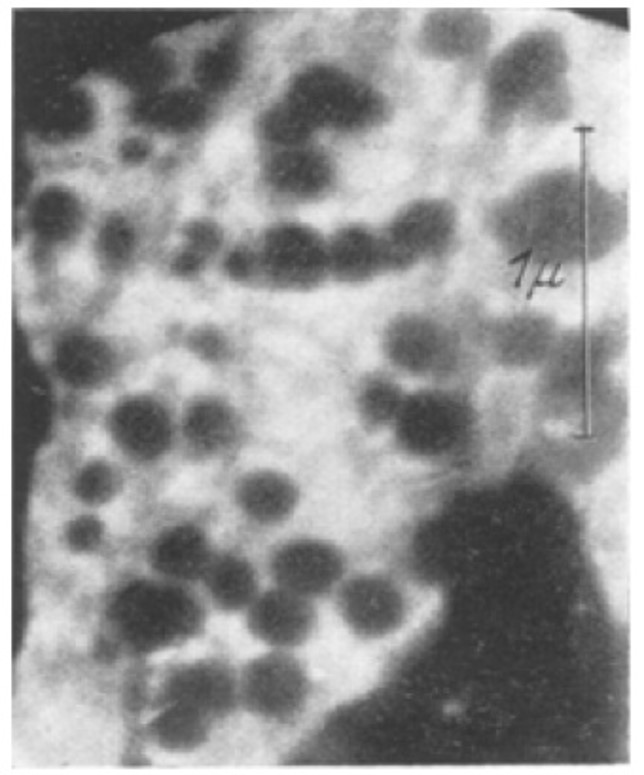
What followed was a golden age for virology as the electron microscope revealed the true nature of different viruses, allowing scientists to understand how viruses operated and how to classify them.
And the new Coronavirus?
Fast forward to 2020: electron microscopy developers were not sleeping. In the 80 years that have passed since the brothers Ruska, many new methods have been developed and existing techniques were refined. And so, after the emergence of a new Coronavirus in the beginning of 2020, it was just a matter of time until EM pictures of SARS-CoV-2 were published. With this we can both see and believe SARS-CoV-2 exists!
Further reading and sources:
Von Borries, B., E. Ruska, and H. Ruska. "Bakterien und Virus in übermikroskopischer Aufnahme." Klinische Wochenschrift 17.27 (1938): 921-925.
Ackermann, Hans-W., and Hans-W. Ackermann. "The first phage electron micrographs." Bacteriophage 1.4 (2011): 225-227.
Knoll, Max, and Ernst Ruska. "Das elektronenmikroskop." Zeitschrift für physik 78.5-6 (1932): 318-339.
Kruger, D. H., Peter Schneck, and H. R. Gelderblom. "Helmut Ruska and the visualisation of viruses." The Lancet 355.9216 (2000): 1713-1717.
Horzinek, Marian C. "The birth of virology." Antonie van Leeuwenhoek 71.1-2 (1997): 15-20.
- *Note that this analogy is not 100% accurate, but for our case this is sufficient.
On Nov 9th, 2020 Pfizer issued a press release stating their conclusion that the COVID-19 vaccine they developed with BioNTech appeared to be 90% effective. While their test contained over 43,000 volunteers they had only detected 94 cases of COVID-19. How confident can you be with only 94 cases? I decided to explore this matter for myself.
I am but a lowly crystallographer, and I’m sure a proper mathematician could do a more rigorous job, but I’ll do the best I can.
The Experimental Design
I was not familiar with the design of this clinical trial but it seems rather straight-forward. You take a whole lot of people and split them into two groups, keeping their group assignment secret from everyone who will be involved in their handling until the end of the trial. The members of one group are given the treatment which we hope is a vaccine while the others are given a sham treatment which is indistinguishable from the “vaccine” by both the participants and their doctors. You then wait to see if anybody comes down with COVID-19.
How long do you wait? You want to wait until you have enough cases to reliably answer the question you hope the study will answer but, to avoid bias, the end point has to be set before the start. If you constantly watch the results and decide to stop when the numbers look good, you could claim success when there is none. After all, life is filled with statistical fluctuations and the results might get worst with longer time.
The press release says that the design of this test says to end after 164 cases of COVID-19 arose among the volunteers but they would peek at the results after 32, 62, and 94 cases. For unspecified reasons they skipped the peek at 32 and, it appears, that the case count shot up to the 94 case trigger while they were discussing the merit of the 62 threshold. I guess this is the only benefit to the world of the huge surge in COVID-19 cases this fall.
It was the 94 case checkpoint that led them to conclude that it was likely that their vaccine candidate was 90% effective at preventing the disease.
But how likely?
To judge the reliability of the 90% number I’ll need to do some statistics. That “proper mathematician” I mentioned earlier would be able to pull out the expected distributions for the experimental results and precisely calculate probabilities and likelihoods. That knowledge is not in my skill set so I’m left with running simulations.
I wrote a program in Mathematica Script to generate many simulations of vaccine trials and then examined their variability. This program has a loop that produces a person with a 100% chance of developing COVID-19 without intervention. That person is assigned either to the Placebo or Vaccinated group. Those poor souls put in the Placebo group are counted as COVID-19 cases. Those in the Vaccinated group are only sickened if they lose a roll of the dice. For each series of simulations I assume a level of effectively for the vaccine. If the run is for a vaccine with 30% effectiveness, for example, the volunteer only get sick if they roll over 30 (okay, I’m using percentile dice.) Those folk protected by the vaccine are let go and the sick are counted as vaccine failures. When the total number of sick reaches the target that trial is complete, the number of sick in each group recorded, and the next trial is started. To ensure that I have a good sample of all possible clinical trials, I simulated a hundred thousand trials for each assumed efficacy.
To keep the numbers simple I ran 100 case trials instead of 94.
When the vaccine is ineffective (can we still call such a thing a vaccine?) there will be an equal number of COVID-19 cases in the Placebo and Vaccinated groups, and this number will be around 50 but there will be variation. If the vaccine is 100% effective the vaccinated group will be completely protected and all 100 cases will be in the Placebo group. The key result of a vaccine trial is the difference in the number of cases. This difference can never be greater than 100 because there aren’t enough cases to result in a bigger number. The difference can, however, be negative since it is possible to have more cases in the Vaccinated group. The most likely explanation for such a result is that the vaccine is very ineffective and randomness of infections happens to result this odd distribution.
Here are my results for a series of hypothetical vaccines with varying efficacy.
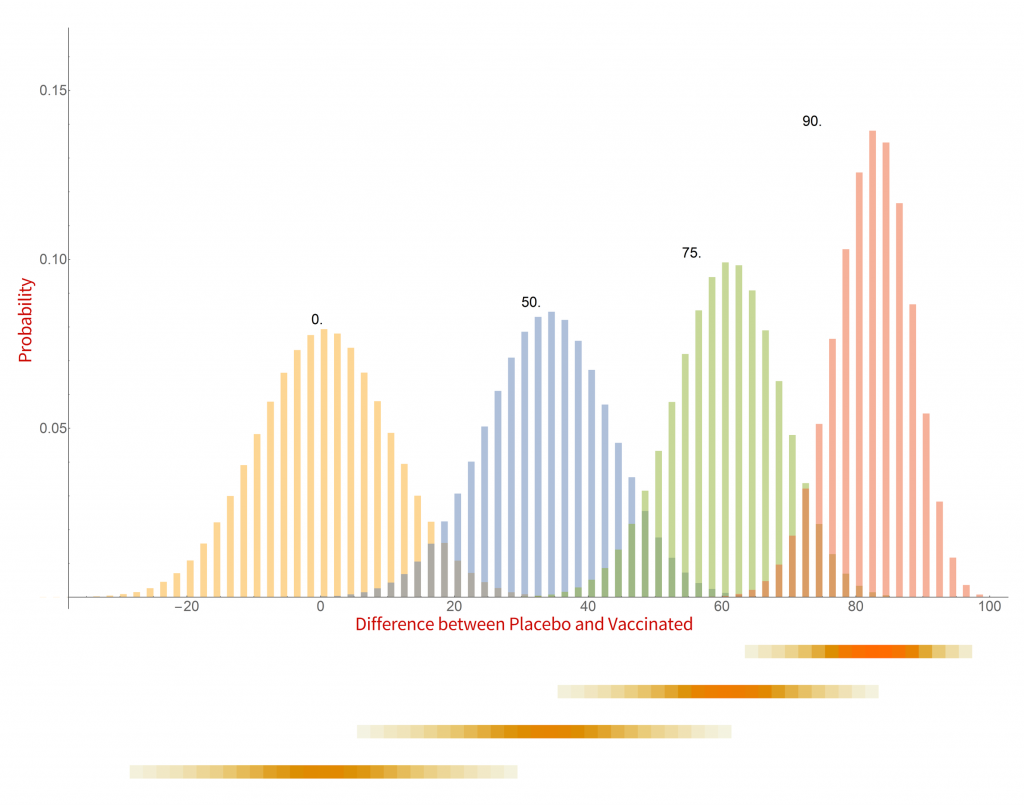
Histograms of the probability of a clinical trial of a vaccine with an assumed efficacy resulting in a particular difference in COVID-19 case numbers between the placebo and vaccinated groups. CC-BY-NC Dale E. Tronrud / Coronavirus Structural Task Force
There are a whole lot of interesting things in this graph. When the vaccine is completely ineffective the most common result of a trial is a difference of zero between the Placebo and Vaccinated groups. There is a fairly wide distribution of results that occur, however. This is the result of statistical fluctuations due to the small number of cases of COVID-19 in the sample (here 100). The distributions for all the simulated efficacies have about the same width, with the exception of those near 100%. Since the difference can never be larger than 100 those distributions get sharper and develop a tail on the lower side.
Let’s look at some scenarios. The graph shows that the most common result of a trial of a vaccine with efficacy of 50% has a difference in number of cases between the Placebo and Vaccinated groups of 36, but sometimes the difference is larger and sometimes smaller. If the vaccine was worthless the most common trial result is zero, but there is also variability. The two histograms overlap considerably which indicates that one cannot distinguish between an efficacy of zero or 50% if the difference in the number of cases in your trial is in the range of zero to about 36. If the difference is greater than this you could conclude that the vaccine is more likely to be 50% than zero percent, and zero percent is the more likely of the two if the difference is negative. Still, there is a wide range of possible outcomes of a trial that have ambiguous interpretation.
On the other hand, what if we have a difference of 80 (90 cases in the Placebo group and 10 in the Vaccinated group)? There isn’t any significant overlap between zero percent efficacy and 90% at the point where the difference is 80. It is much more likely that the vaccine is 90% effective than zero. There is overlap with the 75% effectiveness histogram and we have to admit that it is possible that the vaccine is only 75% effective, but 90% is more likely.
This leads us to realize that the result of a vaccine trial has to result in a range of possible efficacies, with a varying probability of each. My little plot doesn’t make such an assessment very easy. In fact, the plot is starting to show some problems. What it shows is the probability of a trial having a particular result given the effectiveness of the vaccine. What we really want is the probability of each possible efficacy given the result of the clinical trial.
We have to transform our probabilities!
Turning everything on its head
While the calculations I just discussed were easy to set up and understand, they do not really reflect the experiment being done in a vaccine trial. I was assuming an effectiveness of the vaccine and running many, many trials. In reality the effectiveness is unknown and only one trial is run. Where I calculated the probability of a particular difference in COVID-19 cases given the effectiveness of the vaccine what I really want is the probability of the effectiveness of the vaccine given the results of a single clinical trial. It is often difficult to devise such a calculation from scratch but it is pretty straight forward to calculate it from the results I already have.
The first step toward the proper calculation is to expand the current plot. My first figure included simulations of just four possible efficacies. To display more possibilities, I need to abandon histograms. At the bottom of the plot I show four color-shaded bars. In these bars the color is darker when the corresponding histogram is taller. While not as visually clear these bars have the advantage that they can be stacked, and many more plotted in a single figure.
With this new tool I can calculate and display the simulated distribution of clinical trials for every vaccine efficacy from 0% to 100% in 1% steps. The new chart is displayed here.
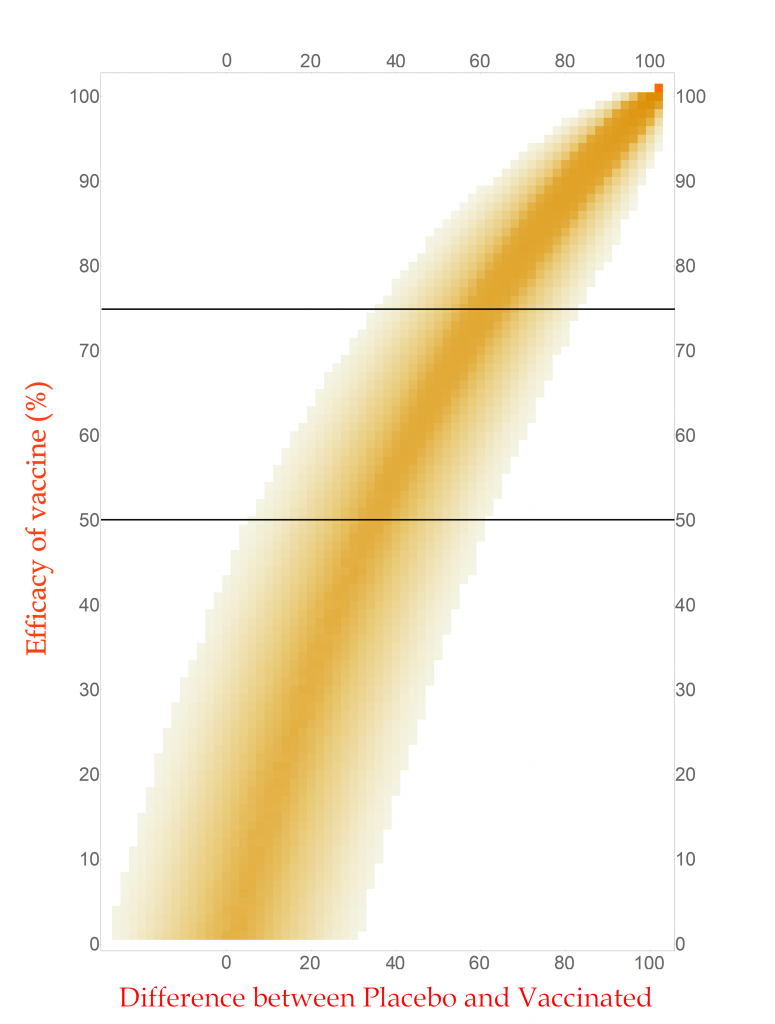
CC-BY-NC Dale E. Tronrud / Coronavirus Structural Task Force
This chart is read by locating the efficacy of your vaccine on the vertical axis and drawing a horizontal line there. The pattern of colors along that line represents the probability of each difference in cases between the Placebo and Vaccinated groups in a clinical trial. I have drawn two such lines, one for a vaccine with 75% effectiveness and another for one with 50%. You can see that the most likely result for the 75% one is about 60 cases (20 in Vaccinated and 80 in Placebo) and the other at about 34 cases. (You figure it out.) In this plot you can see the continuous change as the efficacy of the vaccine is changed. The key point is that there is a spread of results, but I described that before.
With any set of probabilities the full set has to always add up to one. For each horizontal line of colored boxes in this plot the sum of their probabilities is one. A set of numbers with this property is said to be “normalized”.
The vertical lines are not normalized in this plot, as you can see by looking at its left side. There are just a few, very lightly colored or low probability boxes and above them is simply white, which represents zero probability (or at least very, very, very small). This side of the plot has a difference in COVID-19 cases of -24, or in other words the Vaccinated group had 24 more cases of disease than the Placebo. Such an outcome for a clinical trial is very unlikely for any vaccine that has even a tiny amount of success (and is pretty unlikely for one that is merely useless).
Since the probabilities along vertical lines are not normalized they cannot be used as a histogram. Conveniently for us, this can be corrected simply by normalizing them. This is done by summing all the probabilities along each vertical line in this plot and dividing the probabilities in the line by that sum. This gives us a new set of probabilities and a new plot.
How does this magic work? The procedure is justified by a hundreds-of-years-old mathematical theorem called Bayes’s Law. This blog post is already getting long and I leave the application of your favorite search engine to you.
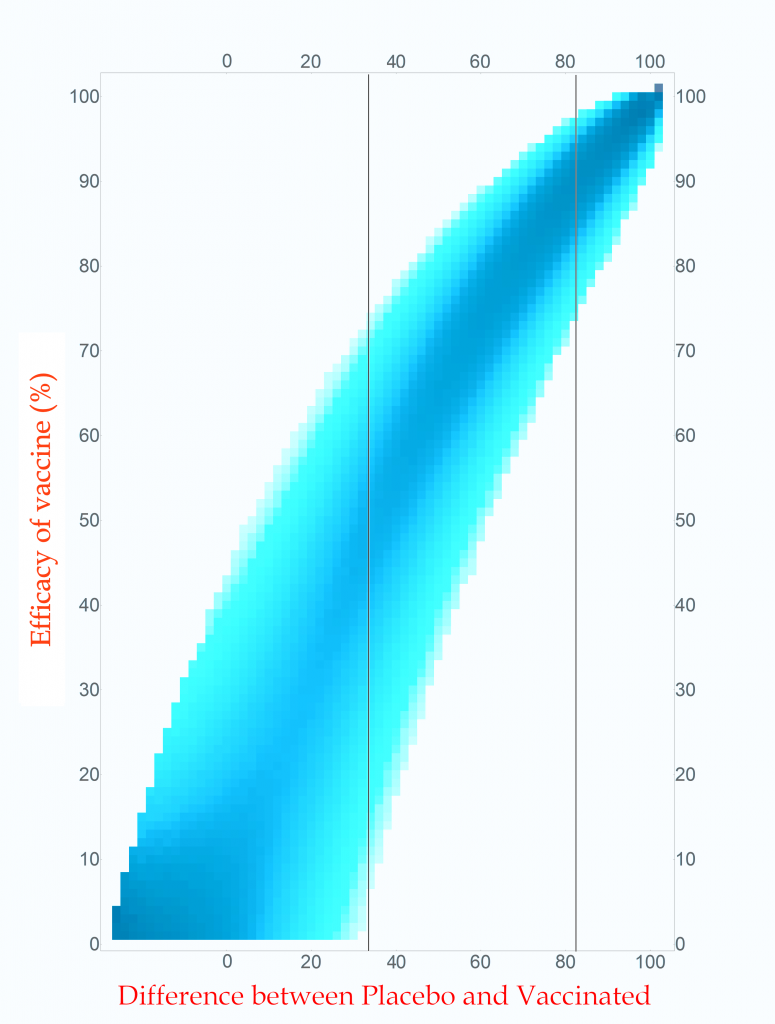
Probability of vaccine effectiveness as a function of clinical trial outcome.
CC-BY-NC Dale E. Tronrud / Coronavirus Structural Task Force
The first thing to note is that the new plot isn’t much different than the original. While the lower-left side has clearly changed, that area is not very interesting. On the right, where the action is, it looks the same. For this reason, many fields of science simply use the unnormalized plot.
The new plot allows us to draw vertical lines (but forbids horizontal lines!). I have drawn example lines at differences of 36 cases and 82 cases. If our clinical trial results in a difference in cases of 82 (91 in Placebo and 9 in Vaccinated) we can see from the line on the plot that the most probable effectiveness of that vaccine is about 90%! This is very close to the happy number reported in Pfizer’s press release. The plot also shows us that there is uncertainty in this number. The vaccine’s effectiveness could be in the mid 70’s or in the upper 90’s.
This is the nature of all experimental work. All results have uncertainties and it is as important to know the amount of uncertainty as it is to know the direct result. You can see how important this is by looking at the 36 case difference line. The darkest blocks along this line, and therefore the most probable efficacy, are near 50%. This would also indicate a useful vaccine, but look at the spread! The width of the uncertainty goes all the way to zero – This vaccine could be worthless. A clinical trial that waited until only 100 cases occurred cannot distinguish between a vaccine with 50% efficacy and a worthless one.
If you wait for more cases to develop the width of the stripe in the plot becomes narrower and the uncertainty drops. The goal of Pfizer was to develop a vaccine of at least 50% efficacy so their design was to wait for 164 cases to give them a narrow enough band to clearly distinguish 50% from zero percent. Just in case the vaccine was better than 50% they built into the design of their trial several points where they could peek and see what was going on. They, and we, lucked out!
What are all those other people for?
The surprising thing about this analysis is that the total number of people in the trial is unimportant when calculating the uncertainty of the result. That answer is the same for a trial with 500 volunteers and a trial with 50,000 volunteers. The only thing that is important is the number of cases of COVID-19.
All those tens of thousands of people are important for other reasons. Very relevant to the current pandemic is that a larger number of volunteers will accumulate the target number of cases sooner: if you double the number of volunteers you will reach the target in half the time. We all want to know as quickly as possible if these vaccines are effective so we want the trials to consist of as many people as the companies can manage.
The other use for large numbers is the search for grim and hopefully rare side effects. These are likely to arise at much lower rates than viral infections and much larger numbers of volunteers are required to achieve statistical significance. A side effect that only occurs in 1 of 5,000 people will require a very large number of participants to be detected. A compounding factor is that a search for an unknown result requires many more data points than the search for a specific outcome, such as COVID-19 infection. (The checks for side effects are only now being described to the public, and I’ll not go into them here.) While the doctors keep an eye out for life threatening problems during the trial, the secret books identifying which volunteers are in which group are kept closed until the end.
End game
After all this interesting math I find that, yes, 94 diseased people are quite enough to conclude that a vaccine is effective at around a 90% level, or at least 70%.
Postscript
During the writing of this post Moderna issued a press release about their vaccine candidate. The analysis presented here applies equally to their trial since I made no assumptions at all about the nature of the vaccine being tested. The only complaint I have with their press release is that they quoted the effectiveness of their vaccine as 94.5%. As you now know, this level of precision is ridiculous. It would be better just to say it is “somewhere around 95% effective”.
The press release
A press release came out today from Pfizer and BioNTech SE with a announcement, you can read the full briefing here.
Suffice it to say leading news outlets all over the world have jumped on the announcement with headlines like “Milestone vaccine offers 90% protection” (www.bbc.com, 9th Nov 2020, 17:00 GMT) or “Covid-19 vaccine candidate is 90% effective, company says” (cnn.com, 9th Nov 2020, 16:00 GMT). But what’s really in the release? Let's have a look!
What does it say?
38,955 people were given two shots. Out of these, half were twice given a real vaccine in development, and half were twice given a placebo. Among all 38,955, 94 people later were proven to have contracted COVID-19: 2,4%. From the cited 90% efficacy, it can be concluded that means: Nine out of approximately 19,478 people who were given the real vaccine contracted COVID-19, while 85 from the placebo-control group did. This may mean the vaccine is effective, leading Pfizer to do this press release. Generally, this is good news. However, one has to consider that the cohort of ill people in total, 94, is still rather small. The press release also explains that the study will finish when 164 patients have contracted COVID-19. This number was likely chosen as they can be pretty certain that a low case number in the vaccine group vs the placebo group was significant at that number of cases and not merely by chance. However, we have a way to go yet: even if the vaccine does prove effective and free of bad side effects, it’s going to be a long while before enough doses can be produced to immunise the whole world. If you want to know about everything that can go wrong in detail, I would like to point you at the legal disclaimer under the press release, which is at least as long as the press release itself and contains beautiful phrases like “risks and uncertainties include (…) the uncertainties inherent in research and development” and “whether and when any such applications may be approved by regulatory authorities, which will depend on myriad factors”.
I am confused
It has to be said that even I, as a scientist, struggled to understand what this press release is saying. Many parts feel confusing, leaving deductions to the reader. For example, 43,538 people were given a first dose, but only 38,955 got a second dose. Were the 4,583 people who only got one dose included in the final results? Apparently not, as they conclude „a vaccine efficacy rate above 90%, at 7 days after the second dose.“
What are “94 evaluable cases” – what constitutes a non-evaluable case?
This becomes only clearer when looking to the clinical trial protocol which is linked from the press release. Here, the evaluable is defined as “no other important protocol deviations as determined by the clinician” and “participants complying with the key protocol criteria” - whatever that means.
What does “approximately 42% of global participants and 30% of U.S. participants have racially and ethnically diverse backgrounds” mean? I am wondering the other 58% / 70% that were not diverse were; I personally suspect that by diverse they meant non-white / non-Caucasian?
Conclusion
The media hype could be a bit more restrained: After all, this is science - but, all in all: Good news with many things that could still go wrong.
____
I would very much like to thank Sam Horrell, Dale Tronrud and Florian Platzmann for discussions and clarifications.
The building plan
Storing the building plans for a virus in its genome is much like how we store ideas in language. This may sound strange but, as an example, typos in spelling, grammar, or word usage, can lead to the meaning of a sentence either changing dramatically, remaining virtually unchanged, or becoming complete nonsense. The SARS-CoV-2 genome consists of RNA. Transcription of this RNA runs into a similar problem: errors can lead to the loss of function, a gain of function, or be completely inconsequential to the resulting protein (Figure 1). Large changes may break the virus, but smaller changes may provide an advantage and are essential for evolution.
Targeting the copy machine
In a previous article we spoke about the copy machinery of the virus, including the RNA-dependent RNA polymerase (RdRp), and drugs targeting it, such as Remdesivir. The goal of these drugs is to jam the enzyme and halt RNA production - or to cause more errors than are sustainable, with the end result being a less infectious virus. The reason the development of drugs targeting the copy machinery of RNA is worthwhile is that humans don’t have machinery to reproduce RNA from RNA. This means drugs targeting this machinery are less likely to interfere with normal processes in people. What if the virus could quickly repair these errors before the new genome is packed into a hull and kicked out the door? That would make finding a therapeutic much more difficult…
Correctional facilities
Unfortunately, SARS-CoV-2 has a way to repair the mistakes. When errors are introduced in transcription through environmental mutagenesis or even mutations caused by nucleotide analogs like Ribavarin1–3, the non-structural protein 14 (nsp14) has the ability to remove them. This multifunctional protein removes errors with the exoribonuclease (ExoN) activity of its N-terminal domain, while the C-terminal domain has the unrelated function of methylating the end cap of the viral RNA3,4.
However, this ExoN does not work alone. There is a replication complex made up of proteins performing many roles in the production of new RNA with high fidelity. Nsp12 is the main hub that makes a new RNA chain to complement the template. Nsp7 and nsp8 have a “processivity” role to enable nsp12 to function efficiently. In addition to these proteins there is a two-component proofreading system of Helicase (nsp13) and the ExoN domain of nsp14. Helicase can detect misshapen RNA helices caused by errors made by the copy machinery5. It then unwinds these double strands of RNA and feeds the strand containing the error into the ExoN domain of nsp14 where they are chopped out. This results in nsp12 continuing RNA replication where it left off.
Exoribonuclease or no exoribonuclease
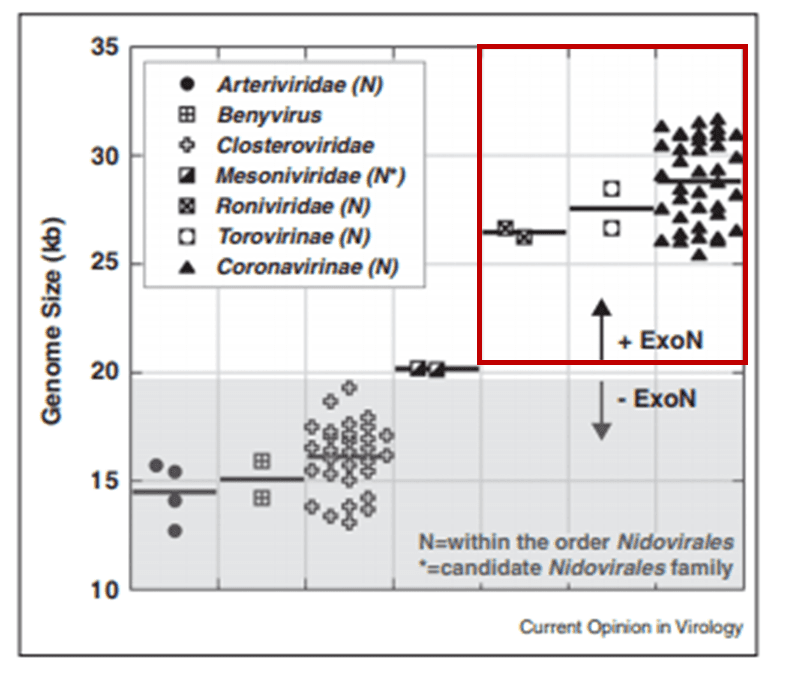
The proofreading ability from Helicase and nsp14 ExoN allows SARS-CoV-2 to have a huge genome as compared to other viruses6(Figure 2). The large 29.9 kb genome of SARS-CoV-2 requires much more physical space to accommodate the necessary genetic information for reproduction when compared to other RNA viruses, such as Rhinovirus that has a genome between 7.2 kb and 8.5 kb in size (Figure 3). When no ExoN proofreading is present genomes cannot expand beyond 20 kb in size6(Figure 2). Maybe by removing the exoribonuclease activity, irreversible damage could be caused to the genome of SARS-CoV-2.

Nsp14 Structure
In order to understand how nsp14 can do this, we need to find out its atomic structure; this may also allow us to develop a drug which hinders its function. However, to this date, no structure of nsp14 from SARS-CoV-2 has been solved. However, structures have been solved of nsp14 in complex with another viral protein, nsp10, both from SARS-CoV (PDB entries 5nfy, 5c8s, 5c8t, 5c8u)2,7. As the protein sequences are very similar between SARS-CoV and SARS-CoV-2 (nsp14 is 95%, and nsp10 is 97% identical), it can be assumed that the SARS-CoV-2 structure as well as its functionality are very similar to SARS-CoV. The active site of the ExoN domain of nsp14 from SARS-CoV-2 has a DEEDh motif (named for the one-letter codes of the amino acids involved) containing a histidine as well as two aspartates and two glutamates2,3,7,8.
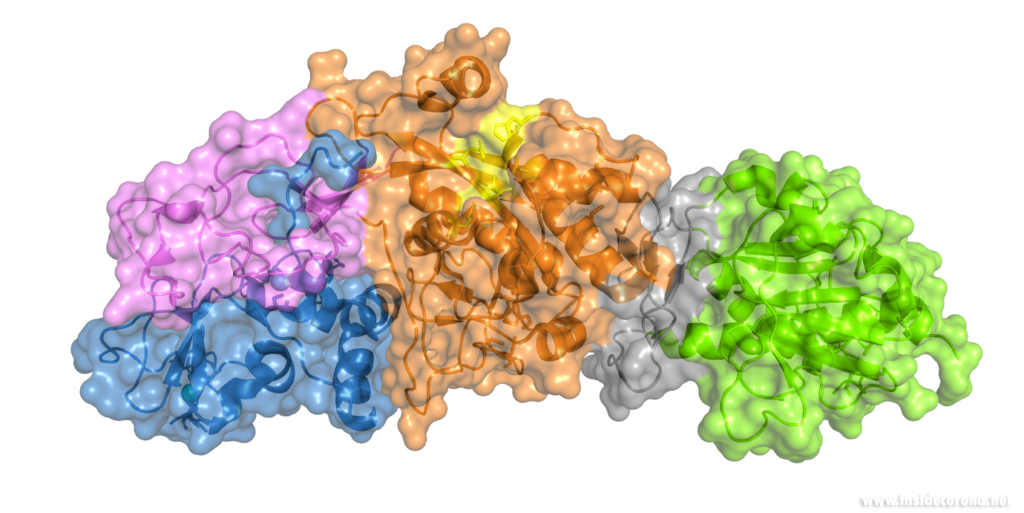
Nsp14 interacts with nsp10
The N-terminus of nsp14 interacts with nsp10 (pink and blue, respectively, in Figure 4). The following domain (orange) has been shown to have exoribonuclease activity on double stranded RNA in a 3’ to 5’ direction9. When nsp10 is interacting with nsp14 there is a 35 fold increase in exoribonuclease activity, which is thought to occur due to conformational changes caused by formation of the complex2,9. The ExoN domain of nsp14 (orange) is connected to the methyltransferase domain (green) by a flexible hinge (black)7,10. This flexible region opens up the methyltransferase active site to allow methylation of the N7 of the 5’ Guanosine triphosphate of RNA10. There are three zinc finger motifs in nsp14 with two found in the ExoN domain and one in the methyltransferase domain2,7. In combination with the two further zinc sites in nsp10, these zinc fingers hold loops of the proteins together and are involved with nucleotide interaction2,7.
Nsp14 has also been demonstrated to form complexes with the copy machinery , nsp12, nsp7, and nsp8, although this interaction is independent of nsp102,11,12.
Exoribonuclease active site and potential drug development
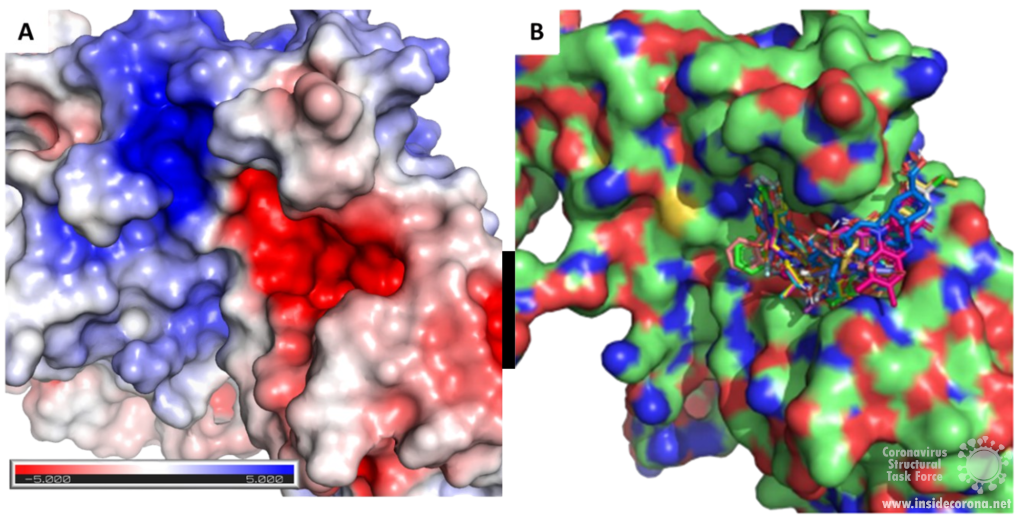
Scientists are searching for drugs that could be used to target nsp14 in order to find a cure for COVID-19. The active site of the ExoN domain of nsp14 has five residues that are essential for activity that form a negatively charged pocket (Figure 5A)7. Currently researchers are using the nsp14 structure from SARS-CoV to model a SARS-CoV-2 structure which can be used to identify compounds that could bind to the active site (Figure 5). These in silico screens start with nucleotide analog drugs like Remdesivir, Ribivarin or Ritonavir that are currently used as antiviral treatments for other viruses13–15. These nucleotide analogs are then changed to achieve a better binding to Nsp14’s active site in order to block it (Figure 5B).
As the ExoN is essential to support the huge 29.9kb genome of SARS-CoV-2, targeting nsp14 could lead to an effective treatment to COVID-19. Although drugs that target just nsp14 could be effective at increasing the error rate in RNA production by the virus, a more effective treatment will require inhibition of the RdRp of the copy machinery at the same time!
Available structures
If you would like to look at the currently available structures for Nsp14(currently only available from SARS-CoV), they are available from our data base; we provide information on the quality of measurement data and models as well as improved structures. The highest resolution structure of nsp14 is PDB entry 5c8t at 3.2Å. This has a bound S-Adenosyl methionine ligand as well as zinc atoms present. Alongside this, another structure of Nsp14 bound to S-Adenosyl homocysteine and a guanosine-triphosphate-adenosine ligand as well as zinc at 3.33Å resolution has been published (PDB: 5c8s). Additionally, two structures with zinc atoms but no ligands are available (PDB 5c8u 3.4Å at and 5nfy at 3.34Å). Both PDB entry 5c8t and 5nfy have improved structures re-refined by our group.
Sources
- 1.Zuo Y. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Research. Published online March 1, 2001:1017-1026. doi:10.1093/nar/29.5.1017
- 2.Ferron F, Subissi L, Silveira De Morais AT, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci USA. Published online December 26, 2017:E162-E171. doi:10.1073/pnas.1718806115
- 3.Barnes MH, Spacciapoli P, Li DH, Brown NC. The 3′–5′ exonuclease site of DNA polymerase III from Gram-positive bacteria: definition of a novel motif structure. Gene. Published online January 1995:45-50. doi:10.1016/0378-1119(95)00530-j
- 4.Chen Y, Cai H, Pan J, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proceedings of the National Academy of Sciences. Published online February 10, 2009:3484-3489. doi:10.1073/pnas.0808790106
- 5.Chen J, Malone B, Llewellyn E, et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Published online July 8, 2020. doi:10.1101/2020.07.08.194084
- 6.Smith EC, Denison MR. Implications of altered replication fidelity on the evolution and pathogenesis of coronaviruses. Current Opinion in Virology. Published online October 2012:519-524. doi:10.1016/j.coviro.2012.07.005
- 7.Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc Natl Acad Sci USA. Published online July 9, 2015:9436-9441. doi:10.1073/pnas.1508686112
- 8.Eckerle LD, Becker MM, Halpin RA, et al. Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing. Emerman M, ed. PLoS Pathog. Published online May 6, 2010:e1000896. doi:10.1371/journal.ppat.1000896
- 9.Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proceedings of the National Academy of Sciences. Published online May 25, 2012:9372-9377. doi:10.1073/pnas.1201130109
- 10.Ogando NS, Ferron F, Decroly E, Canard B, Posthuma CC, Snijder EJ. The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity. Front Microbiol. Published online August 7, 2019. doi:10.3389/fmicb.2019.01813
- 11.Subissi L, Posthuma CC, Collet A, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA. Published online September 2, 2014:E3900-E3909. doi:10.1073/pnas.1323705111
- 12.Subissi L, Imbert I, Ferron F, et al. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: Replicative enzymes as antiviral targets. Antiviral Research. Published online January 2014:122-130. doi:10.1016/j.antiviral.2013.11.006
- 13.Khater S, Dasgupta N, Das G. Combining SARS-CoV-2 proofreading exonuclease and RNA-dependent RNA polymerase inhibitors as a strategy to combat COVID-19: a high-throughput in silico screen. Published online June 24, 2020. doi:10.31219/osf.io/7x5ek
- 14.Shannon A, Le NT-T, Selisko B, et al. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Research. Published online June 2020:104793. doi:10.1016/j.antiviral.2020.104793
- 15.Narayanan N, Nair DT. Ritonavir May Inhibit Exoribonuclease Activity of Nsp14 from the SARS-CoV-2 Virus and Potentiate the Activity of Chain Terminating Drugs. chemrxiv.org. Published May 13, 2020. https://chemrxiv.org/articles/Ritonavir_May_Inhibit_Exoribonuclease_Activity_of_Nsp14_from_the_SARS-CoV-2_Virus_and_Potentiate_the_Activity_of_Chain_Terminating_Drugs/12280043
Introduction
Have you heard that the coronavirus “mutates”? Or that there are “several strains” of it around the world? Sounds scary, right? However, the reality is that everything “mutates”. All organisms, over time, acquire differences in their genes, from bacteria to humans. You might be aware that this can happen when your DNA (Deoxyribonucleic Acid) is exposed to UV light (like from the sun!), but this can also happen during DNA replication. This is when a cell uses the template of one of the two DNA strands to make a new complimentary copy of the other strand. Mutation is common to all living organisms (and viruses) and a driver of evolution. This is the first post in a series that will explore coronavirus replication with a focus on the proteins involved.
How does the coronavirus make more of itself?
SARS-CoV-2 uses single-strand Ribonucleic acid (RNA) to encode its genome, not DNA, and hence belongs to a class of “single-strand RNA viruses”. For this reason, the virus needs a different way to copy its genome than “normal” cells have. The viral protein that copies the RNA is called an “RNA-dependent RNA polymerase” (RdRp). This protein uses the viral RNA as a template to make a new copy of viral RNA, by stringing single ribonucleotides together like beads on a string. This process is called polymerization.
A study by the Morse lab at Texas A&M University showed that SARS-CoV-2 RNA polymerase has a remarkable similarity to the RNA polymerase of SARS-CoV (>95%) as well as MERS-CoV [1], the virus which causes Middle-Eastern Respiratory Syndrome. This means that research performed in response to the SARS and MERS epidemics can inform our response to SARS-CoV-2. Unfortunately, a lack of consistent pandemic-preparedness funding means that we didn’t learn as much about RdRp in time as we could have. Still, RNA polymerase might be a viable drug target for halting the spread and reducing the fatality rate of COVID-19.
Structure of the RNA-Dependent RNA Polymerase
By determining the structure of RdRp, and deeply understanding how it works, we can optimize a drug to specifically target it and hinder its function. To this end, in the last few months, several structures of SARS-CoV-2 RNA polymerase have been published.
One interesting structure shows RNA polymerase in action, in the process of elongating an RNA strand (see Figure 1).[2] This structure clearly show the polymerase in complex with smaller proteins, non-structural protein 7 and 8 (nsp7 and nsp8). These proteins improve how well the RNA polymerase binds the template RNA and also how long it stays bound before dissociating – a feature called “processivity”.[3]
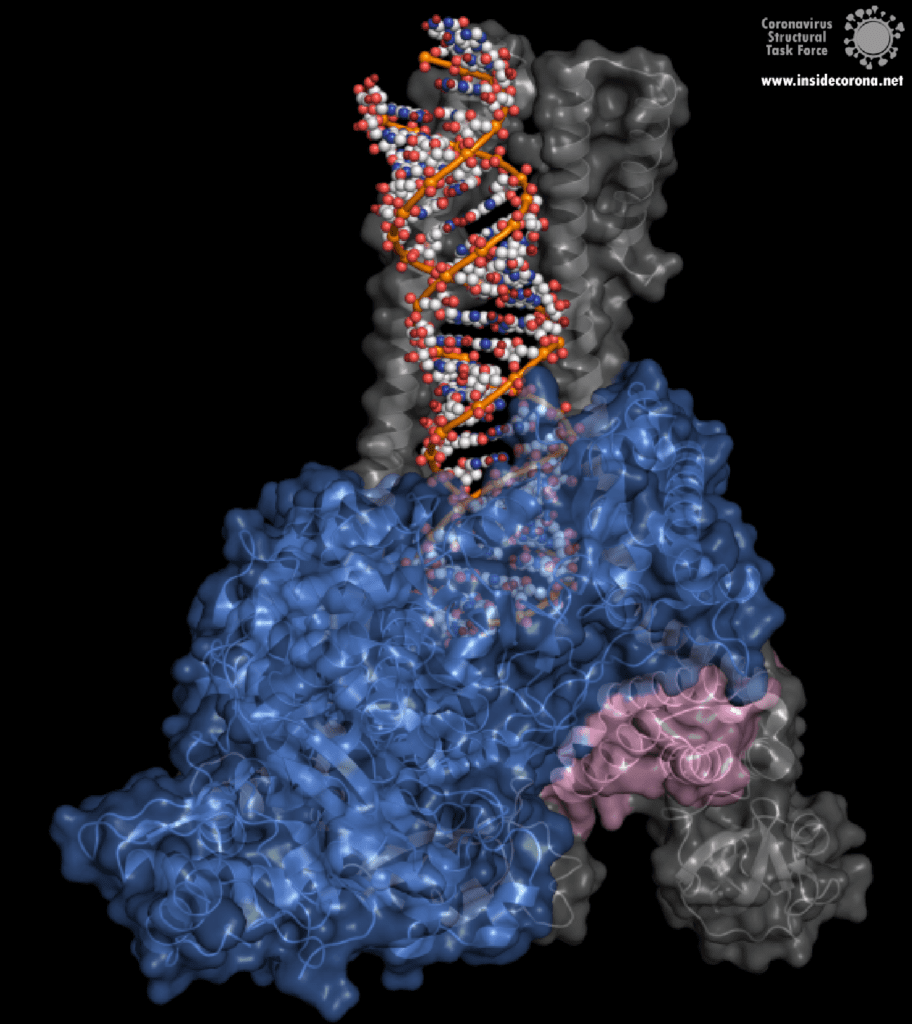
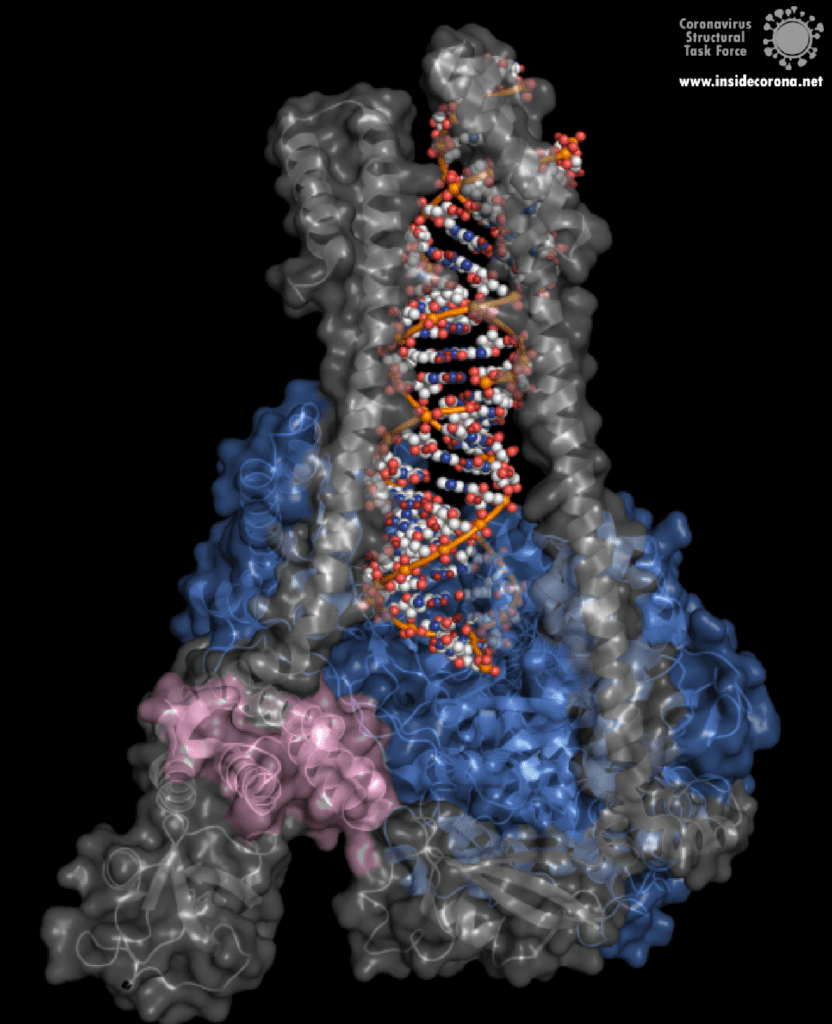
In the center of the protein is the area where the main action happens, called the “active site”. The amino acids of the polymerase that form the active site have a particular shape and chemical properties, which enable the polymerization reaction to occur very rapidly. In fact, the polymerase can string together as many as 100 nucleotides per second! [3] New RNA molecules can enter the active site through a little window to be added to the growing RNA chain. It is here that the antiviral drugs make their move!
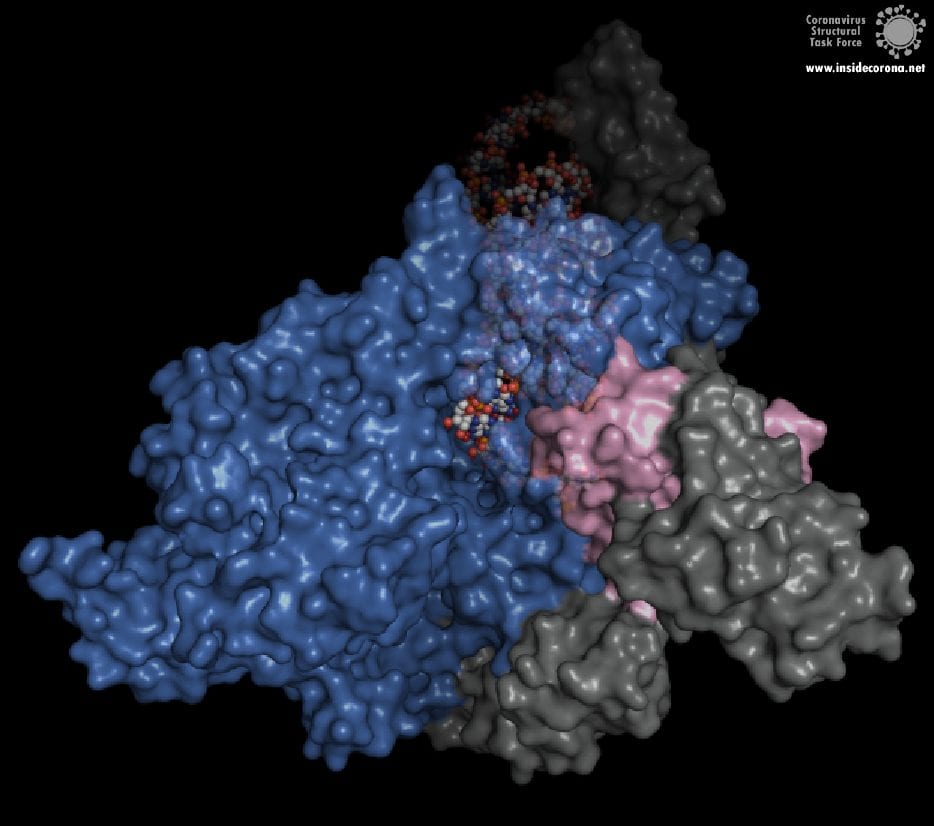
How do antiviral drugs attack RNA-dependent RNA polymerase?
First, let’s talk about Gilead’s FDA-approved drug, Remdesivir, which has taken the spotlight in the search for COVID-19 cures. Remdesivir (which has a fancy chemistry ID, GS-5734, and is sold under the brand name Veklury), is a “nucleotide analog”, which means that it mimics the shape and chemistry of the nucleotides that make up RNA and DNA (see figure).
Remdesivir was developed originally as a general antiviral drug and was later shown to protect cells (in a test tube) and monkeys (not in a test tube) from the Ebola Virus [4]. However, this was recent enough, and science is slow enough that, until the COVID-19 pandemic, large-scale clinical trials of Remdesivir hadn’t been done yet. So scientists and doctors have been rushing to test the drug in COVID-19 patients. In fact, the US and Japan both approved the drug for “Emergency Use Authorization'' for severe COVID-19 patients as early as May [5], [6]. And, in July, the European Medicines Agency gave Remdesivir a “conditional marketing authorization” (used for drugs that meet an unmet medical need but have insufficient data for normal approval). This allows the use of Remdesivir in severe COVID-19 patients through the next year [7]. So, how the heck does a drug for Ebola, Influenza, or some other viruses also work against COVID-19? I was concerned by this when the news about all the drug trials were coming out – and I’m sure I wasn’t the only one...
The simple answer to that is all these viruses need to do the same thing - copy their RNA genome from an RNA template. And in order to do that, they all end up using basically the same tool, an RNA-Dependent RNA polymerase. And all drugs that are nucleotide analogs use the very same trick: they dress up like ribonucleotides (the "beads on a string" from before) and fool the RNA polymerase into letting them into the active site. Once inside, they get “stuck” in the active site, jamming the polymerase machine. Since this trick should work for any viral RNA polymerase, we can use these drugs for any RNA virus, and call them ‘general antivirals’. Of course, in practice, this doesn't always work, because there are differences between the different RNA polymerases. However, it is a great place to start! In the future, if we have general antivirals for SARS-CoV-2 all ready-to-go, we may be better equipped to deal with another coronavirus outbreak!

The Chemistry of Remdesivir
Remdesivir resembles the nucleotide adenine in structure, although it has some fancy chemical add-ons which help make it a better drug (thank you, medicinal chemistry!). When Remdesivir is injected into a vein, it travels through the bloodstream and enters into our cells, which recognize it as a foreign substance and try to digest it. However, what ends up happening is that the cells remove just the fancy chemical add-ons, and then confuse it for a normal adenine nucleotide. In infected cells, the viral RNA-dependent RNA polymerase then starts grabbing these molecules and inserting them into the new viral RNA strand in place of adenine molecules. Remdesivir, now attached to the RNA, jams the polymerase, rendering the virus unable to make more copies of its genome. Ultimately, this halts viral replication and helps the patient fight off the virus.
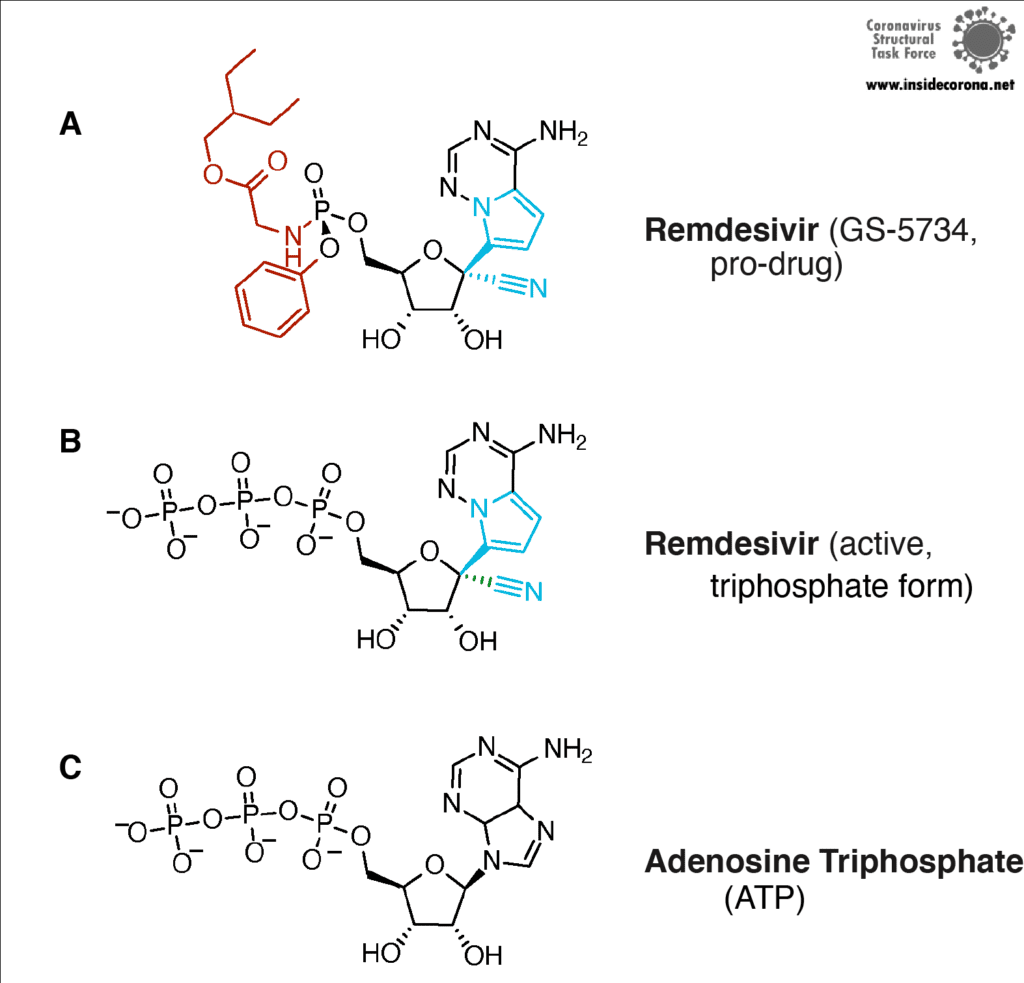
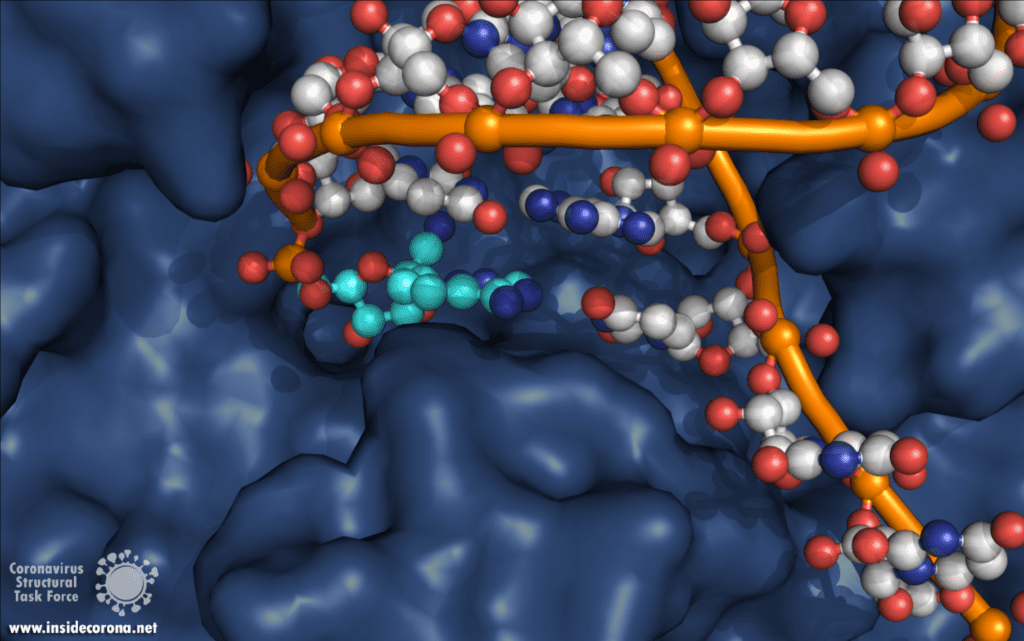
Another drug that inhibits the RNA polymerase activity is Favipiravir, sold under brand names Avigan, Abigan, and FabiFlu. Favipiravir has been discovered by Toyama Chemical Co., Ltd. in Japan and it has a similar mechanism to Remdesivir, except that it mimics a guanosine nucleoside instead of an adenine nucleotide [8]. This drug was approved in Japan back in 2014 for use in resistant cases of Influenza A and B, but still remains unapproved in the US (still in Phase II and Phase III clinical trials) and the UK [9]. This drug is also being tested for use against Ebola virus, Lassa virus, and currently SARS-CoV-2 in 43 countries. The approval of Favipiravir for COVID-19 has been much faster in China (Mar 15, 2020), Russia (Jun 3, 2020), and India (Jun 20, 2020)[10], [11]. Nonetheless, other countries, including Japan, are in various stages of clinical trials, and the results are anticipated to be out by the end of July [10].
So...do we have a cure for SARS-CoV-2?
Sadly, not yet. While the speed at which Remdesivir has gone through clinical trials is unprecedented, more work needs to be done to make sure it is safe and effective. Since (in the big scope of things) not a lot have people have taken Remdesivir, we aren’t really sure what all the side effects are, although there is emerging evidence for liver and kidney damage [12, 13]. The most common side effects are nausea (10% and 9% of patients), indigestion (7%) and increase of transaminases (6% and 8%). In one study, 3.6% of patients in a 10-day trial needed to stop taking therapy due to the latter. However, serious viral infections can also cause liver damage, so separating the two causes is a challenge! Remdesivir is not a cure-all, either. In one study it improved the recovery time from 15 days to 11 days, but it showed no effect for patients with mild to moderate disease, and no difference in median recovery time for patients who were already on a ventilator [14]. Since the drug has to be given by infusion over several days, there is a pretty small window in which Remdesivir can actually help.
Likewise, Favipiravir has its own side effects such as liver damage, elevated uric acid levels, kidney damage, skin allergies, etc. [15]. These effects restrict it for use by severe diabetes and heart patients. On top of that, it is not suitable for pregnant women because it can cause potential fetal deaths and deformities. It has been shown that Favipiravir works only during the earlier stages of SARS-CoV-2 infection when the body’s immune system isn’t totally drained, whereas it can result in a cytokine storm (when your immune system really freaks out) in severely ill patients. But, unfortunately, the virus doesn’t differentiate between humans while attacking, so a universal drug for COVID-19 has to be safe for use by all people.
However, these drugs are better than nothing, and by understanding the mechanisms involved, scientists can continue to improve upon the existing drugs for the benefit of all. While most of the ‘general antivirals’ that target RNA Polymerase have failed with SARS-CoV-2, Remdesivir has been relatively successful. Scientists think that this is actually because of a proofreading protein in SARS-CoV-2 called exonuclease. Immediately after the RNA-polymerase makes new RNA, exnuclease checks to make sure the new RNA is correct. In one study, another drug that mimics RNA called Ribivarin was shown to be removed from newly synthesized RNA by exonuclease [16]. Thankfully, Remdesivir is not excised , which is likely why it has been more successful than the other options [17], [18]. To read more about how nsp14 maintains the integrity and virulence of SARS-CoV-2, tune in to a future blog entry!
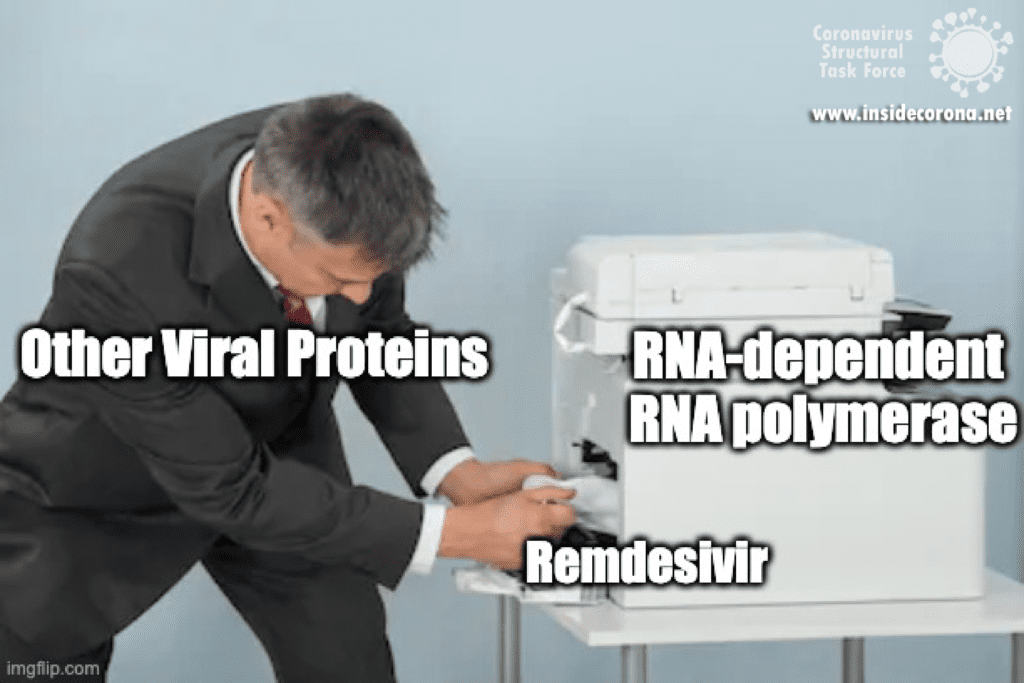
Recommended Structures
For those interested in reviewing the structures further, they are available in our GitHub repo, along with information about validation and, where relevant, improved structures. For a high-resolution comparison of the active site with and without Remdesivir, 7BV2 and 7BV1 (respectively) were published together at 2.5 and 2.8 Å. The elongating structure of the complex shown above (6YYT) has the polymerase as well as the cofactors and RNA very well resolved, with little "missing" density and a resolution of 2.9 Å. It is likely preferable to 6M71 and 7BTF, which were published with a similar resolution but with less of the complex resolved, and no RNA. For those interested, 7C2K and 7BZF (at 2.93 Å and 3.26 Å) show the complex bound to RNA in a pre- and post-translocation state.
Sources
[1] J. S. Morse, T. Lalonde, S. Xu, and W. R. Liu, “Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV,” ChemBioChem, vol. 21, no. 5, pp. 730–738, Mar. 2020, doi: 10.1002/cbic.202000047.
[2] H. S. Hillen, G. Kokic, L. Farnung, C. Dienemann, D. Tegunov, and P. Cramer, “Structure of replicating SARS-CoV-2 polymerase,” Nature, May 2020, doi: 10.1038/s41586-020-2368-8.
[3] W. Yin et al., “Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir,” Science, p. eabc1560, May 2020, doi: 10.1126/science.abc1560.
[4] R. T. Eastman et al., “Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19,” ACS Cent. Sci., May 2020, doi: 10.1021/acscentsci.0c00489.
[5] O. of the Commissioner, “Coronavirus (COVID-19) Update: FDA Issues Emergency Use Authorization for Potential COVID-19 Treatment,” FDA, May 04, 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-emergency-use-authorization-potential-covid-19-treatment (accessed Jul. 08, 2020).
[6] A. Sternlicht, “Japan Approves Remdesivir For Use On Severe COVID-19 Patients,” Forbes. https://www.forbes.com/sites/alexandrasternlicht/2020/05/07/japan-approves-remdesivir-for-use-on-severe-covid-19-patients/ (accessed Jul. 08, 2020).
[7] D. CZARSKA-THORLEY, “First COVID-19 treatment recommended for EU authorisation,” European Medicines Agency, Jun. 25, 2020. https://www.ema.europa.eu/en/news/first-covid-19-treatment-recommended-eu-authorisation (accessed Jul. 10, 2020).
[8] E. De Clercq, “New Nucleoside Analogues for the Treatment of Hemorrhagic Fever Virus Infections,” Chem. Asian J., vol. 14, no. 22, pp. 3962–3968, Nov. 2019, doi: 10.1002/asia.201900841.
[9] K. Shiraki and T. Daikoku, “Favipiravir, an anti-influenza drug against life-threatening RNA virus infections,” Pharmacol. Ther., vol. 209, p. 107512, May 2020, doi: 10.1016/j.pharmthera.2020.107512.
[10] T. Hornyak, “Japan sending Fujifilm’s flu drug favipiravir to over 40 countries for Covid-19 trials,” CNBC, May 04, 2020. https://www.cnbc.com/2020/05/04/fujifilms-flu-drug-favipiravir-sent-to-43-nations-for-covid-19-trials.html (accessed Jul. 14, 2020).
[11] G. P. Ltd, “Glenmark Becomes the First Pharmaceutical Company in India to Receive Regulatory Approval for Oral Antiviral Favipiravir, for the Treatment of Mild to Moderate COVID-19.” https://www.prnewswire.com/in/news-releases/glenmark-becomes-the-first-pharmaceutical-company-in-india-to-receive-regulatory-approval-for-oral-antiviral-favipiravir-for-the-treatment-of-mild-to-moderate-covid-19-855346546.html (accessed Jul. 14, 2020).
[12] Goldman, J. D. et al. Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. N. Engl. J. Med. (2020) doi:10.1056/NEJMoa2015301
[13] Remdesivir Safety Forecast: Watch the Liver, Kidneys | MedPage Today. https://www.medpagetoday.com/infectiousdisease/covid19/86582
[14] J. H. Beigel et al., “Remdesivir for the Treatment of Covid-19 — Preliminary Report,” N. Engl. J. Med., vol. 0, no. 0, p. null, May 2020, doi: 10.1056/NEJMoa2007764.
[15] Sandhya Ramesh, “Favipiravir, Japanese drug that’s the new Covid treatment hope your chemist will soon stock,” ThePrint, Jun. 25, 2020. https://theprint.in/health/favipiravir-japanese-drug-thats-the-new-covid-treatment-hope-your-chemist-will-soon-stock/447987/ (accessed Jul. 14, 2020).
[16] F. Ferron et al., “Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA,” Proc. Natl. Acad. Sci., vol. 115, no. 2, pp. E162–E171, Jan. 2018, doi: 10.1073/pnas.1718806115.
[17] C. J. Gordon, E. P. Tchesnokov, J. Y. Feng, D. P. Porter, and M. Gotte, “The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus,” J. Biol. Chem., Feb. 2020, doi: 10.1074/jbc.AC120.013056.
[18] L. Zhang et al., “Role of 1’-Ribose Cyano Substitution for Remdesivir to Effectively Inhibit both Nucleotide Addition and Proofreading in SARS-CoV-2 Viral RNA Replication,” bioRxiv, p. 2020.04.27.063859, Apr. 2020, doi: 10.1101/2020.04.27.063859.
The coronavirus cannot be seen with the naked eye; it is invisible. That is a huge problem. Imagine if your house were on fire: you would react immediately, leave the house, call the fire brigade and warn the neighbours. The thread would be clearly visible. This is, however, not true for the coronavirus. SARS-CoV-2 cannot be seen or touched. The time between infection and tangible illness is a number of days, with an additional 16 days until the worst-case scenario, death (2). This makes the threat much harder to recognize. Imagine a catastrophe killing 32.362 people in New York City! Still, the same number of deaths occured in recent month in NY – 1 in 250 persons – by COVID-19 (3). The invisibility of the threat makes it hard for people to wear masks and keep distance to each other. It is very difficult to adress a danger you can’t see, and even harder to believe in something you don’t understand.
This is why we want people to see the virus, or even better, to touch it. Make it tangible. And this is why we have designed a scientifically accurate model for 3D print. And the best part: As we are funded by taxpayer money, we have made the files available online for free! (If you use them, please acknowledge the Coronavirus Structural Task Force.)

Now that is a bit different from what you have seen in the media, isn’t it? Why? Well, first of all, any kind of spikey ball passes as a corona virus these days. Our virus also differs from the illustration by medical illustrators Alissa Eckert and Dan Higgins from the CDC (4). These are the main differences between their red-and-grey depiction and ours:
- The virus is not exactly spherical, and rather wobbly; which can be seen clearly in our 3D print.
- The number of Envelope proteins (orange), membrane proteins (yellow) and spikes (green) are in accordance to the latest findings (2).
- The spikes are longer, as we now know much more about their structure, and the virus hull is smaller in proportion to them. However, the exact size of the virus varies. The ratio we show is an average.
- The spikes are glycosylated, making them more irregular (and slimier). The glycosylation is shown in grey. However, it would actually look like this, and could be represented by cotton wool stuck to the spike protein.
- We are showing the E protein in the pentameric “pore” conformation. Whether that is a correct assumption for the virion remains to be seen. If you want to know more about this, look here.
- As the virus lives embedded in slimy, wet conditions, we chose colours to represent that instead of the red and grey which the CDC used to represent the threat the virus poses (1)
We have also added a rhinovirus and an antibody in the same 1 : 1,000,000 scale. The antibody binds to the spike specifically, this is how the immune system can recognize the virus – and the rhinovirus shows you how large SARS-CoV-2 is. (Spoilers, it is very big for a virus!)
One should also say that the "virus" is actually a virion - the transport form of a virus, which can be spread - once a host cell is infected, the virus rolls out his RNA genome and makes a whole lot more proteins to hijack the host cell and make more copies of itself. These proteins are also part of the virus, and a main subject of our research.
References:
(1) Rothan Hussin A., and Byrareddy Siddappa N. "The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak." Journal of autoimmunity (2020): 102433. pmid:32113704
(2) https://wwwnc.cdc.gov/eid/article/26/7/20-0282_article
(3) 32.362 deaths by 17th of July 2020 as reported by the New York Times in 8.4 million inhabitants (US Census Bureau) gives 0,39 %, roughly equalling 1 in 250.
(4) https://www.nytimes.com/2020/04/01/health/coronavirus-illustration-cdc.html
