David C. Briggs, Luise Kandler, Lisa Schmidt, Gianluca Santoni & Andrea
Thorn
This blog post was published in Crystallography Reviews.
Please cite: https://doi.org/10.1080/0889311X.2023.2173744
Abstract
The coronavirus SARS-CoV-2 is the causative agent for the COVID-19 pandemic. Its proteome is typically separated into three classes of proteins: (1) Structural proteins which facilitate the transport and host cell infiltration of the viral RNA, (2) non-structural proteins which are thought to be essential for the viral life cycle and are all produced from open reading frame 1ab (ORF1ab) on the RNA, and (3) everything else, called accessory proteins. Although it was originally thought that these accessory proteins are non-essential for viral replication, a growing body of evidence suggests that these diverse proteins have crucial roles in virus-host interactions, in particular in the way they interfere with the signalling pathways that modulate the host cell’s response to infection and viral pathogenicity. Here, we summarize efforts to structurally characterize the accessory proteins from SARS-CoV-2.
Introduction
Synthesis of accessory proteins
During host infection, SARS-CoV-2 viral particles bind to the ACE2 receptor on the surface of host cells via their Spike protein. Following entry of the virion into the cell, (either via membrane fusion or endocytosis), the virus sheds its coat and ORF1ab of the positive sense RNA viral genome is translated into protein by host ribosomes. The proteases within the ORF1ab transcript cleave the polyprotein into the Non-structural proteins. A subset of these proteins, nsps 7,8,9,10,12,13,14, and 16, come together to form the replication and translation complex (RTC). The RTC is responsible for initiation of RNA replication, and subgenomic RNA (sgRNA) synthesis. It is these subgenomic RNAs that serve as the template for accessory protein synthesis.
Subgenomic RNAs are synthesized from the 3’ end of the genomic RNA. Synthesis continues until a transcriptional regulatory sequences (TRS) is encountered, at which point, the replication and translation complex may pause and jump to another, complementary transcriptional regulatory sequences. The negative sense transcript is transferred to the complementary leader transcriptional regulatory sequences (through base-pairing interactions) and transcription continues until the RTC reaches the 5’ end of the genome.
The skipping of certain sequences due to the transcriptional regulatory sequences leads to a range of product sizes, all sharing the same leader sequence. Once transcription is complete, the negative sense RNA template now serves as a template for positive strand synthesis, again performed by the replication and translation complex. In turn, the various positive sense subgenomic RNAs produced serve as templates for viralmRNA production, resulting in expression of proteins not encoded by the large polyprotein [1]. Additionally, full-length, positive sense genomic RNA is packaged into newly produced viral particles [2].
Table 1. Summary of SARS-CoV-2 accessory proteins with function and available structures.
| Accessory protein | Function | Length (aa) | Known host binding partners | PDB entries | References |
| ORF3a | Ion channel, Autophagy inhibitor. | 275 | 6XDC 7KJR | [4] | |
| ORF6 | Interferon antagonist | 61 | mRNA export factor RAE-1, Nup98 | 7VPH | [5] |
| ORF7a | Inhibits host defenses that prevent virion release | 121 | Tetherin | 6W37 7CI3 | No publication [6] |
| ORF7b | Unknown – golgi resident | 43 | N/A | ||
| ORF8 | Down regulates MHC I expression & interferon response | 121 | IL17RA receptor, Spike-protein | 7JTL 7JX6 7F5F | [7] no publication [8] |
| ORF9b | Disrupts Interferon response | 97 | TOM70 | 6Z4U∗ 7DHG 7KDT | No publication [9, 10] |
Location in the SARS-CoV-2 genome
The accessory proteins are six small to medium-sized proteins with a length between 43 and 275 amino acid residues (Table 1). Unlike many of the non-structural proteins that are encoded within the large polyprotein ORF1ab (which is expressed as one long polyprotein chain and proteolytically cleaved to yield individual proteins), accessory proteins are each encoded by their own open reading frames (ORFs). In SARS-CoV-2, these ORFs are found at the 3’ end of the SARS-CoV-2 genome between the structural proteins, or in the case of ORF9b, within an alternative reading frame of the nucleocapsid gene (Figure 1(A)). ORF3a (275 amino acids long) is found eight nucleotides after the end of the gene encoding the Spike protein and before that the envelope protein. ORF6 (61 amino acids), ORF7a (121 amino acids), ORF7b (43 amino acids) and ORF8 (121 amino acids) are located between the genes for the membrane protein and nucleocapsid. The gene for accessory protein ORF9b (97 amino acids) is found within an alternative open reading frame1 ten bases after the start codon for the nucleocapsid gene.
This review covers SARS-CoV-2 accessory proteins as described in Uniprot [3] as having ‘evidence at a protein level’. Other accessory proteins have been proposed to be produced during SARS-CoV-2 infection, however, evidence for their expression is uncertain at the time of writing. This ambiguity exists because while open reading frames are detected in the SARS-CoV-2 genome, evidence of protein expression during infection of host cells (either by western blot or mass spectrometry) is lacking.
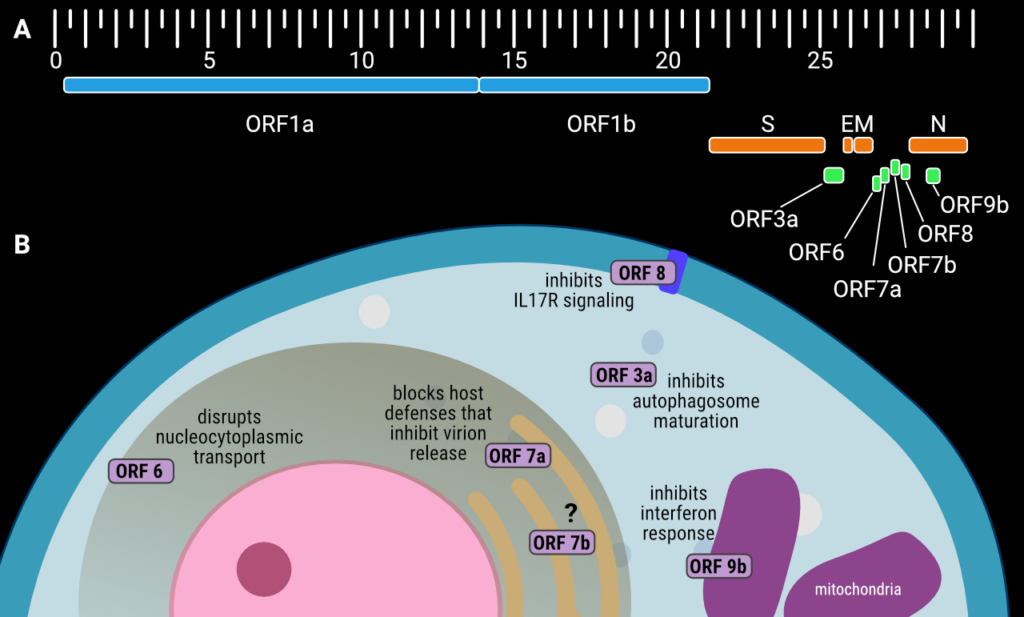
Figure 1. (A) The genetic organization of the SARS-CoV-2 genome, with accessory proteins highlighted in green. The scale is in kilobase pairs, positions are approximate, (B) schematic showing the subcellular localization and function of the SARS-CoV-2 accessory proteins during host cell infection. Creator: Coronavirus Structural Task Force - Lisa Schmidt, License: cc-by-sa
Physiological roles of SARS-CoV-2 accessory proteins
Cell-based assays reveal that whilst the accessory proteins are not essential for viral replication, SARS-CoV-2 variants lacking ORF6, ORF7a, and ORF8 have altered viral replication kinetics. This means slower production of progeny virions, or less efficient virion release or reinfection following production in the host cell [11]. The same study shows that in humanized, transgenic mice SARS-CoV-2 infection with ORF3a-deficient viruses and, perhaps to a lesser extent, ORF6-deficient viruses, results in reduced lethality compared to wild type SARS-CoV-2 infection.
ORF3a
SARS-CoV-2 ORF3a is a dimeric viroporin, an integral membrane protein that functions as a non-selective, Ca2+-permeable ion channel [4]. Despite classification as an accessory protein, it does contribute to efficient replication and viral release. By binding to VPS39, a protein involved in vesicle sorting, SARS-CoV-2 ORF3a blocks the fusion of autophagosomes with lysosomes hence inhibiting the maturation of the autophagosomes (vesicles where unwanted material such as damaged organelles and invading pathogens are isolated prior to degradation) and thereby hindering autophagy as a cellular defence mechanism.
This binding mechanism blocks the assembly of the STX17–SNAP29–VAMP8 SNARE complex – a protein complex required for the fusion of autophagosomes and lysosomes, a crucial process by which the hydrolytic enzymes present in the lysosomes are released into the autophagosomes to help break down unwanted materials including invading pathogens which have been isolated for degradation [12].
In addition to this, it has been shown that SARS-CoV-1 ORF3a activates the NLRP3 inflammasome. NLRP3 (NACHT, LRR and PYD domains-containing protein 3) is an intracellular sensor that becomes activated on detection of pathogens or other danger signals.
This activation triggers release of pro-inflammatory cytokines, Interleukins (IL) -1B and – 18 [13]. SARS-CoV-1 ORF3a has also been shown to promote expression of IL-1B, via an interaction with TRAF3 (Tumour necrosis factor receptor–associated factor 3). This interaction with TRAF3 promotes the ubiquitination of ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain) and p105 (Nuclear factor NF-kappa-B p105 subunit). Ubiquitination of ASC results in the activation of caspase 1, a protease required for IL-1B and IL-18 maturation [14]. Upon ubiquitination p105 yields the p50 protein; this promotes the NF-kappa-B proinflammatory response, which drives pathologic inflammation. SARS-CoV-2 ORF3a retains the SARS-CoV-1 TRAF binding motif (SARS-CoV-1: 36−PLQAS−40, SARS-CoV-2: 36−PIQAS−40) and therefore is assumed to retain this function, identified and characterized in [15].
ORF6
ORF6 is a multifunctional disruptor of interferon (IFN) signalling and antiviral immunity. It has been shown to accomplish this by interrupting nucleocytoplasmic transport through direct interaction with the Ribonucleic acid export 1 (RAE1) – Nuclear pore complex protein 98 (NUP98) protein complex [16,17]. This interaction downregulates IFN-Beta production by preventing Interferon regulatory factor 3 (IRF3) from moving into the nucleus.
An interaction with Karyopherin α 2 blocks ORF6 from importing IRF3 into the nucleus [18]. Once in the nucleus, IRF3 binds to the interferon-stimulated response element, and activates transcription of INF-1 and -2 and associated IFN-stimulated genes as part of the host response to infection [19]. The inhibition of NUP98 also prevents interferon signalling by blocking Signal Transducer and Activator of Transcription 1 (STAT1) entering the nucleus [20].
ORF6 also inactivates the Major Histocompatibility Complex (MHC) class I pathway, by inhibiting NLR Family CARD Domain Containing protein 5 (NLRC5), a key activator in host antiviral response [21]. Recent studies have shown that ORF6-mediated cytotoxicity is inhibited by the selective nuclear export inhibitor Selinexor (but not Ivermectin) [22], and this could formthe basis for future drug discovery projects to ameliorate SARS-CoV-2 symptoms.
ORF7a
SARS-CoV-2 ORF7a is a single-pass type I membrane protein thought to be retained on the surface of the endoplasmic reticulum (ER) after expression into the ER lumen, where it antagonizes the Tetherin protein (also called Bone marrow stromal antigen 2 (BST2)) [23]. Tetherin is a host defense protein that tethers nascent virions to the host membrane, preventing their dispersal. As such inhibition of Tetherin function aids virion release and subsequent infection of host cells. ORF7a can also become polyubiquitinated with Lys63-linked ubiquitin on Lys-119, located on the cytoplasmic side of the ER membrane. This modification leads to antagonism of IFN-1 signalling by blocking Signal Transducer and Activator of Transcription 2 (STAT2) phosphorylation [24].
ORF7b
Very little is known aboutORF7b, but in SARS-CoV-1 (which bears ∼81% sequence identity with SARS-CoV-2 ORF7b), it is found inside the Golgi compartment of the host cell and is has also been shown to be present in viral particles [25]. Expression of SARS-CoV-2 ORF7b within HEK-293 or Vero E6 cells induces expression of a range of cytokines including IFN-β, Tumour necrosis factor alpha (TNF-α) and IL-6, and activated type-I IFN signalling via phosphorylation of IRF3. Ultimately, ORF7b activates TNFα-induced apoptosis [26]. Proteomic analysis suggests that ORF7b might interact with innate immunity regulators Mitochondrial antiviral-signalling protein (MAVS) and Unc-93 homolog B1 (UNC93B1) [27].
ORF8
ORF8 is a multifunctional protein encoded in a hypervariable region of the SARS-CoV-2 genome [28] and is not conserved in SARS-CoV-1. ORF8 protein has a signal peptide and is secreted outside the cell [29], where it interacts with and activates host Interleukin 17 receptor alpha (IL17RA) by mimicking the natural ligand, Interleukin-17A (IL-17A) [30]. This interaction ultimately leads to the activation of NF-KB and expression of pro-inflammatory cytokines, contributing to the SARS-CoV-2 cytokine storm [31]. It has also been demonstrated that ORF8 interacts directly with the S1 region SARS-CoV-2 Spike protein, and downregulates Spike expression and S1/S2 cleavage, perhaps explaining the observation that variants lacking ORF8 have increased transmissibility [32]. In addition to this, ORF8 downregulates MHC-I levels on the cell surface by directly binding to MHC-I and promoting its degradation [33]. This downregulation results in SARS-CoV-2-infected cells being less susceptible to cytotoxic T-lymphocyte-mediated lysis. ORF8 can also function inside the cell as a histonemimic as it contains an ‘ARKS’ motif, located in a disordered loop in the crystal structure. This ARKS motif allows ORF8 to interact with chromatin and various histone-modifying enzymes resulting in the alteration of transcription of genes associated with response to viral infection [34]. It should be noted that much of the ORF8 protein in variant B.1.1.7 (Alpha Variant) is missing due to a Q27STOPmutation [35] but the full-length protein is found in subsequent variants. There is also a polymorphism that results in a L84S mutation – the S84 variant is associated with milder disease and less severe clinical outcomes [36].
ORF9b
ORF9b is a multi-functional and structurally dimorphic protein encoded within an alternative ORF within the SARS-CoV-2 nucleocapsid protein. In SARS-CoV-1, it is expressed through leaky ribosome scanning of the nucleocapsid mRNA [37]. SARS-CoV-2 ORF9b directly interacts with the mitochondrial import receptor subunit TOM70. This binding inhibits activation of an antiviral signalling cascade by locking the TOM70 in a state which has impaired binding to the Heat shock protein 90 (Hsp90) Glu-Glu-Val-Asp (EEVD) motif [10]. The interaction between TOM70 and Hsp90 is important for importing proteins from the cytosol into the mitochondria, which in turn is essential for mitochondria function [38]. Curiously however, the full-length ORF9b protein is incapable of binding to TOM70 [9]. The mature ORF9b protein exists as a dimer with a hydrophobic central cavity assumed to be involved in lipid binding (see later) and is consistent with a role in virion maturation (PDB: 6Z4U).
Structural overview of SARS-CoV-2 accessory proteins
Structures have so far only been solved for SARS-CoV-2 ORF3a (PDB entries 7KJR, 6XDC), ORF 6 (PDB entry 7VPH), ORF7a (PDB entries 7CI3, 6W37), ORF8 (PDB entries 7JTL, 7JX6, 7F5F) and ORF9b (PDB entries 7DHG, 6Z4U, 7KDT). No structures exist at the time of writing for ORF7b.
ORF3a
The cryo-EM structure of SARS-CoV-2 ORF3a has been determined with a single ORF3a homodimer contained within a lipid nanodisc [4]. The PDB contains two structures, 6XDC at 2.9Å resolution and 7KJR at 2.08Å resolution, both determined by the same research group. The model 7KJR is the higher resolution structure, and so all discussion of the ORF3a structure will focus on this. The maps are of excellent quality and show two ordered molecules of the lipid phosphatidylethanolamine, as well as visible interactions with the MSP1E3D1 scaffold protein (a nanodisc component).
As can be seen in Figure 2(A), ORF3a consists of an N-terminus (amino acids 1–39), a C-terminus (amino acids 239–275) and a short cytoplasmic loop (amino acids 175–180). The transmembrane domain of each symmetrically composed protomer consists of three helices. Together, the six transmembrane helices forman elliptical ion channel in the membrane. The pore is oriented with its N-terminus on the luminal side and the C-terminus on the cytosolic side. The ORF3a viroporin possesses a novel fold, with low structural homology to any other protein structure of the PDB. The third transmembrane helix of each protomer is followed by a helix-turn-helix motif connecting the transmembrane domain with the C-terminal cytosolic domain. An eight-stranded β-sheet sandwich forms a hydrophobic core of this cytosolic domain and links the two monomers through close Van der Waals interactions between V168, V225, F230 and I232.
There are several polar cavities within the ion channel, which are thought to reveal the path of the transiting ions. Structural studies of different conformational states may be required to ascertain whether this is correct [4]. Kern and colleagues were also able to isolate anddetermine the structure of a tetrameric form of ORF3a, albeit at lower resolution (6.5 Å). This tetramer is composed of two dimers side-by-side, however, it is currently not known if this assembly plays a physiological role (see EMDB entry EMD-22138).
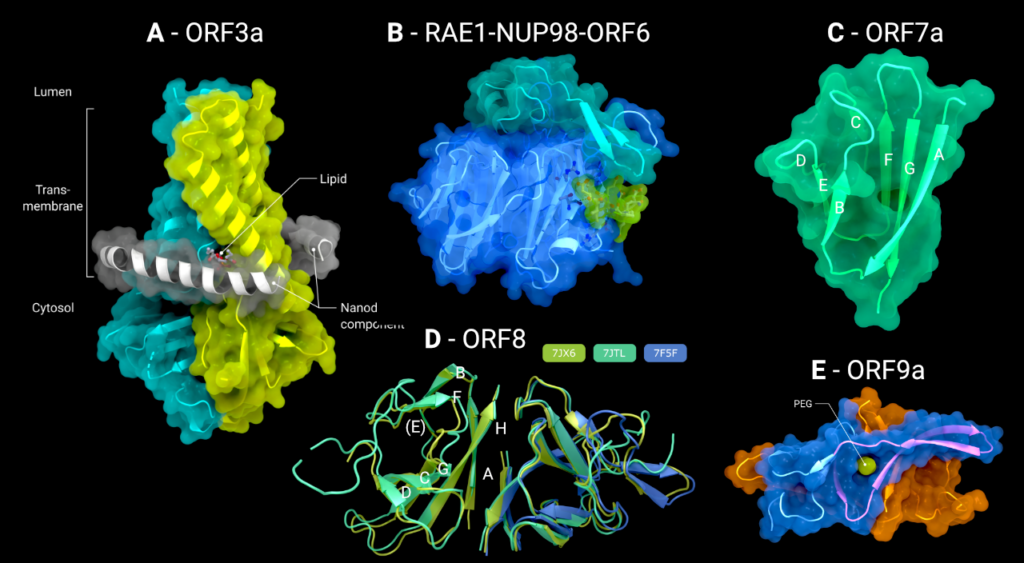
Figure 2. (A) The cryo-EM structure of the viroporin ORF3a (PDB: 7KJR) in a lipid nanodisc (ordered nanodisc components in grey), (B) the crystal structure of the tail of ORF6 (green), bound to the RAE1–Nup98 (blue/cyan) nucleoporin pair (PDB: 7VPH), (C) the crystal structure of the luminal domain of ORF7a (PDB: 7CI3), (D) the crystal structure of the disulphide-linked dimer of ORF8 (PDB: 7J6X, 7JTL, 7F5F), (E) the crystal structure of the full-length ORF9a dimer with bound PEG/Lipid molecule (green) (PDB: 6Z4U). Creator: Coronavirus Structural Task Force - Lisa Schmidt, License: cc-by-sa
ORF6
ORF6 is a small protein of only 61 amino acid residues. Secondary structure predictions with Jpred [39] and structure prediction with Colabfold implementation of Alphafold2 [40,41] suggest the protein is composed almost entirely of a single α-helix with a small unstructured carboxy-terminus (Colabfold models available at 10.5281/zenodo.7323979). The only structural information for ORF6 comes from a structure of the ternary complex formed from a synthesized peptide corresponding to residues 41 to 61 of ORF6 with the mRNA export factors Nup98 and RAE-1. In this structure, only the ORF6 C-terminal sequence DEEQPMEID (residues 53–61) is ordered and binds to the outer face of blades 5 and 6 of the seven-bladed β-propeller of RAE-1 (Figure 2(B)). The surface patch of the RAE1-Nup98 complex occupied by ORF6 is positively charged, and is thought to be the mRNA binding site that RAE1-Nup98 uses to export host mRNA from the nucleus to the cytosol, thus preventing this interaction inhibits host protein synthesis [5].
ORF7a
The N-terminal domain of ORF7a consists of seven β-strands arranged in an immunoglobulin (IG) like β-sandwich fold with greatest structural similarity to the Intercellular adhesion molecule (ICAM) IG-fold (Figure 2(C)). Sequence analysis of SARS-CoV-1 predicts that ORF7a encodes for a type I transmembrane protein with 122 amino acid residues, including a 15-amino-acid-long signal peptide at the N-terminus, a single transmembrane helix of ∼21 amino acid residues, and an endoplasmic reticulum retention signal at the C-terminus. These conclusions would suggest that the N-terminal IG-like domain would be present in the lumen of the endoplasmic reticulum, and indeed, subcellular localization studies confirm this [22].
The structure of the IG domain of ORF7a has been determined twice, both times by X-ray crystallography. Structure 6W37 covers residues 16–82, was determined to 2.9Å resolution and as yet has no accompanying publication. Structure 7CI3 is composed of residues 14–96 and was determined to 2.2Å resolution [6].
Within the folded domain, the IG-fold has seven β-strands ordered in two β-sheets consisting of four β-strands (A, G, F, C) in the first sheet and three (B, E, D) in the second one. Both sheets are amphipathic, with the hydrophobic side facing inwards closely packed against each other. The top of the ectodomain is defined by the BC, DE and FG loops and the bottom by the AB, CD and EF loops. The β-sandwich structure is stabilized by two disulphide bonds linking the sheets at opposite edges. At the bottom of the structure, a disulphide bridge connects Cys23 on strandAwithCys58 at the end of strandE. At the top, Cys35 of the BC loop is linked to Cys67 at the end of strand F (Figure 2(C)). Additionally, on top of the BED sheet, the DE loop protrudes from the structure and forms a groove together with β-strands C andD. In the centre is Glu33, which contributes to the negatively charged bottom of the mainly hydrophobic groove. This groove may be a potential site for ligand or cation interaction due to its central negative electrostatic potential.
ORF7b
ORF7b is a short 43 amino acid residues long protein, and as yet no structures exist of any portion of it. Despite the lack of a predicted signal sequence [42], it is thought that the protein is incorporated into the membrane of the Golgi apparatus and the produced viral particles, as has been demonstrated for SARS-CoV-1 [25]. Like ORF6, ORF7b is predicted to be formed almost entirely of a single alpha-helix, with a disordered carboxy-terminus (Colabfold models available at 10.5281/zenodo.7323984).
ORF8
ORF8 is secreted from the infected cell into the extracellular space. It has an N-terminal signal sequence, followed by a single domain of roughly 106 amino acids. There are three Xray crystal structures of SARS-CoV-2 ORF8 in the PDB: 7JTL, which was determined to a resolution of 2.04Å [7], 7JX6, which was determined to 1.6Å (no associated publication), and 7F5F also determined to 1.6Å [8] (Figure 2(D)). It is worth noting that all proteins were expressed in E. coli and hence lack the N-linked glycosylation on Asn78 [29]. There exists some intrinsic flexibility within the (physiologically relevant) dimer, as can be seen from the subtle changes in orientation in the right-hand monomer in Figure 3(A), when the three dimers are optimally superimposed on the left-hand monomer.
In all three structures, ORF8 forms a disulphide-linked homodimer through Cys20 (although in PDB 7F5F, the dimer spans two asymmetric units), and each protomer contains three additional intramolecular disulphide bonds. The fold of each protomer resembles an immunoglobulin fold, with β-strands B, E (not a β-strand in all structures) & F forming one sheet and strands C, D, G & H forming another sheet. Strand A also forms a β-sheet with strand H, and this two-stranded sheet makes up much of the hydrophobic homodimerization interface. A long and partially disordered loop exists between strands D & E.
There is a polymorphism in ORF8 where the amino acid at position 84 is switched from a hydrophobic leucine to a hydrophilic serine amino acid residue; here the site of the L84S polymorphism is on a face of the ORF8 structure that also contains the N78 Nlinked glycosylation site. A recent preprint suggests that ORF8 can be secreted through an unconventional YIF1B-mediated pathway, where it bypasses the host cell glycosylation machinery in the endoplasmic reticulum [43]. Lin et al demonstrate that this unglycosylated ORF8 is the form that binds to, and activates, IL17RA, rather than the glycosylated from. Taken alongside the L84S polymorphism, this finding allows us to speculate that the IL17RA binding site is on this face of ORF8 that carries both N78 and L84 (Figure 3(B)).
Clearly, further research is necessary to structurally characterize this interaction – if it turns out that this interaction might be amenable to disruption by small molecules or monoclonal antibodies, either by targeting ORF8 or IL17RA [31], therapeutics might be developed to prevent the debilitating cytokine storm, caused by ORF8, as seen in some COVID-19 patients.
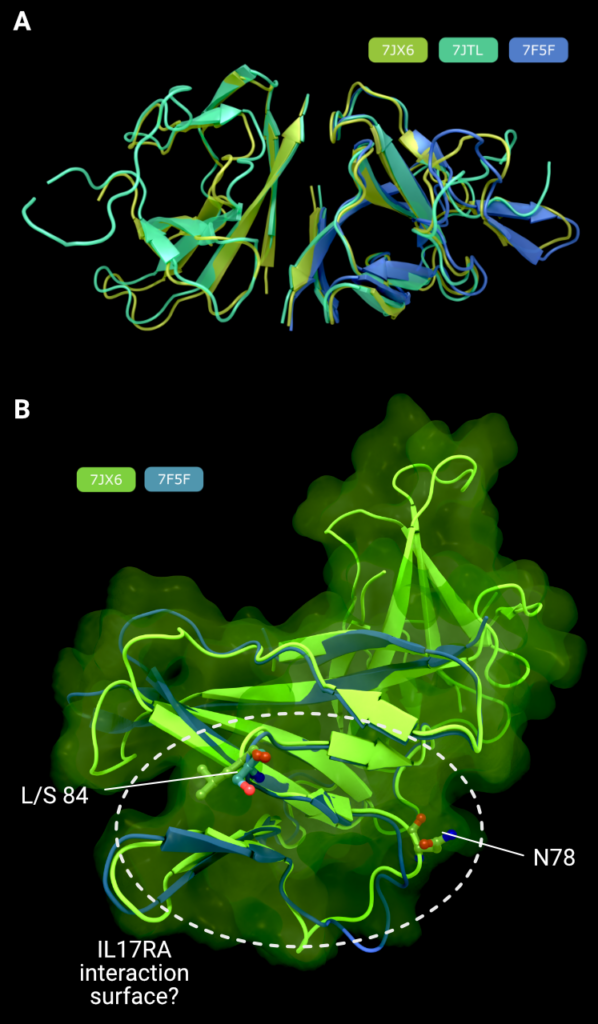
Figure 3. (A) Comparison of all three structures of SARS-CoV-2 ORF8, superimposed on the right-hand monomer. (B) Comparison of the L84 (PDB: 7JX6, green) and S84 (PDB: 7F5F, blue) variants of ORF8 showing the putative IL17RA binding surface that harbours the both polymorphism at position 84, and the N-linked glycosylation site at position 78, known to inhibit IL17RA binding in glycosylated ORF8. Creator: Coronavirus Structural Task Force - Lisa Schmidt, License: cc-by-sa
ORF9b
There are three structures of ORF9b in the PDB. The most complete structure is PDB entry 6Z4U, which contains residues 1 through 97 and has no accompanying publication. The data go to 1.95Å resolution and were phased by molecular replacement from the SARSCoV-1 ORF9b structure, 2CME [44]. The maps and the model-to-map fit are generally of good quality. The two structures share the same fold, with an RMSD of 0.91Å over 64 matched atom pairs. ORF9b exists as a highly interconnected dimer in solution, with an all-β topology that resembles a pair of β-barrels with extensive strand swapping and extended loops (Figure 2(E)). A hydrophobic cavity can be found in between the two protomers.
In the SARS-CoV-1 structure, this cavity has an extended fatty acyl chainmodelled into it, consistent with a role as a lipid-binding protein [44]. In the SARS-CoV-2 structure, the cavity has been modelled holding a polyethylene glycol chain from the crystallization solution. Examination of the electron density maps suggest that either solution could be correct, as both proteins were crystallized from a PEG-containing crystallization buffer, although Meier et al. did conduct mass spectrometry experiments to confirm the presence of a long chain fatty acid in the SARS-CoV-1 structure. The absence of an accompanying publication for the SARS-CoV-2 structure hinders certain identification of the bound hydrophobic ligand in PDB entry 6Z4U.
The remaining two structures of ORF9b show a C-terminal peptide of ORF9b (residues 43-78) in complex with the mitochondrial import receptor, TOM70 (Figure 4(B)). PDB 7KDT [10] (3.05Å cryo-EM structure) and 7DHG [9] (2.2Å X-ray crystallographic structure) are similar with an RMSD of 1.1Å over 416 C-alpha pairs. The structures contain residues 39–78 (7KDT) and 43–78 (7DHG) of ORF9b, which adopt a long α-helix (residues 51–70) with the N-terminus of the helix interacting with a cleft in the C-terminal TOM70 tetratricopeptide repeat (TPR) domain. PISA [45] analysis shows an interaction area of ∼2000Å2 between TOM70 and the Orf9b C-terminal peptide. The central portion of the bound helix (residues 58–66) has predominantly electrostatic interactions with TOM70, whereas the N-terminal end of the helix (residue 45–54) are more hydrophobic in nature, with several aliphatic side chains docking into non-polar pockets of TOM70 (Figure 4(C)).
Gao et al. report that the affinity of the C-peptide for TOM70 as KD = 0.96 μM, which is ∼2.6 times tighter than the interaction with the Hsp90 EEVD peptide. Interestingly, although they are predicted to bind at separate sites, pre-incubating TOM70 with ORF9b C-peptide reduced the affinity of TOM70 for EEVD by almost 30-fold (KD = 72.99 μM).
Taken together, these data suggest ORF9b is an allosteric inhibitor of the interaction between TOM70 and Hsp90, and it perhaps does this by locking the TOM70 in a conformation that is less able to bind Hsp90 – this results in the inhibition of downstream interferon activation.
Curiously, this TOM70-bound structure is completely incompatible with the lipidbound, all beta-sheet, full length form determined in PDB entry 6Z4U(Figure 4(A,B)), and indeed Gao et al. report that the full length ORF9b protein does not interact with TOM70 and only a truncated version or an alpha-helical peptide corresponding to the region seen in 7KDT and 7DHG bind to TOM70. It is not yet known what causes this switch between the two forms of ORF9b. This could happen at a transcript or translation level, or as the result of a proteolytic event, or perhaps that the all-β ORF9b structure observed in 6Z4U unfolds in the absence of a lipid, allowing the unfolded protein to adopt the α-helical conformation observed in the TOM70-bound forms. Further research is needed to determine the mechanism(s) that control this switch. Despite the unanswered questions regarding ORF9b, it is conceivable that a small molecule could be developed to prevent the interaction of TOM70 and ORF9b, and that this might alleviate symptoms by preventing SARS-CoV-2 from circumventing host anti-viral responses.
Summary
So far, efforts to structurally characterize the accessory proteins of SARS-CoV-2 have been largely successful, only ORF6 and ORF7b lack experimentally determined structures for most of the sequence. Given thatmuchof these twoproteins are thought tobe unstructured or just comprise a single alpha helix, it is not clear how beneficial isolated structures of these proteins will be to investigate their biological role. Structures in complex with host proteins (where known), however, would provide molecular details of interactions that may be targets for future drug design efforts. Despite their characterization as ‘accessory’, many of these proteins do have roles that promote effective and efficient infection. For example, ORF3a has been shown to promote efficient release of new viral particles, and ORF8 may regulate Spike protein presentation and maturation. Whilst they may not be high-priority for drug screening and design, it is clear that inhibition of the function of accessory proteins (e.g. blocking ORF8 IL17RA interactions) might serve to alleviate the symptoms of SARS-CoV-2 infection in vulnerable patients.
Despite their structural diversity, the SARS-CoV-2 accessory proteins primarily serve to block host cell reactions to viral infection. The interferon signalling pathway, a critical element in the activation of anti-viral defense, is inhibited by ORF6 (preventing transport of IRF3 into the nucleus to active transcription of interferon-response genes) and ORF7a (through blocking of STAT2 activation). Pro-inflammatory cytokine production and maturation (which can lead or contribute to the cytokine storm) are upregulated by ORF3a, ORF6, ORF7b and ORF8. Counterintuitively, whilst inflammation is an immune response to pathogens, over stimulation of the immune system can be beneficial to pathogens, as is seen in superantigens, proteins that subvert the hosts immune systemby over activating it, causing indiscriminate and off-target damage to host tissues [46]. ORF3a also inhibits host defenses by blocking the degradation of invading viruses by phagocytosis.
In terms of future work on the accessory proteins, the lack of data about ORF6 should be addressed once a binding partner is found. In addition to this, whilst we have multiple structures of ORF8, structures of this protein in complex with either Interleukin 17 Receptor A, or with SARS-CoV-2 spike protein would be most instructive, and either of these interactions, particularly the one with IL17RA, may prove to be a target for future therapeutic design efforts.
This blog post was published in Crystallography Reviews, please cite: https://doi.org/10.1080/0889311X.2023.2173744
Acknowledgements
The authors would also like to thank Johannes Kaub and Rosemary Wilson for support and discussion. Figures in this review use the Protein Imager interface [47].
Funding
This work was supported by the German Federal Ministry of Education and Research [grant number 05K19WWA and 05K22GU5] and Deutsche Forschungsgemeinschaft [project TH2135/2-1]. D.C.B. acknowledges that this work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK [grant number CC2068], the UK Medical Research Council [grant number CC2068] and the Wellcome Trust [grant number CC2068].
References
[1] Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. Epub 2009/05/12.
[2] Hartenian E, Nandakumar D, Lari A, et al. The molecular virology of coronaviruses. J Biol Chem. 2020;295:12910–12934. Epub 2020/07/15.
[3] UniProt C. Uniprot: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D4D9. Epub 2020/11/26.
[4] Kern DM, Sorum B, Mali SS, et al. Cryo-EM structure of the SARS-CoV-2 3a ion channel in lipid nanodiscs. bioRxiv. 2021. Epub 2020/06/27.
[5] Li T, Wen Y, Guo H, et al. Molecular mechanism of SARS-CoVs Orf6 targeting the Rae1-Nup98 complex to competewithmRNAnuclear export. FrontMol Biosci. 2021;8:813248. Epub 2022/02/01.
[6] Zhou Z, Huang C, Zhou Z, et al. Structural insight reveals SARS-CoV-2 ORF7a as an immunomodulating factor for human CD14(+) monocytes. iScience. 2021;24:102187. Epub 2021/02/23.
[7] Flower TG, Buffalo CZ, Hooy RM, et al. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc Natl Acad Sci U S A. 2021;118(2):e2021785118. Epub 2020/12/29.
[8] Chen X, Zhou Z, Huang C, et al. Crystal structures of bat and human coronavirusORF8 protein Ig-like domain provide insights into the diversity of immune responses. Front Immunol. 2021;12:807134. Epub 2022/01/04.
[9] Gao X, Zhu K, Qin B, et al. Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions. Nat Commun. 2021;12:2843. Epub 2021/05/16.
[10] Gordon DE, Hiatt J, Bouhaddou M, et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370:eabe9403. Epub 2020/ 10/17.
[11] Silvas JA, Vasquez DM, Park JG, et al. Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 human ACE2 transgenic mice. J Virol. 2021;95:e0040221. Epub 2021/06/17.
[12] Miao G, Zhao H, Li Y, et al. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev Cell. 2021;56:427–442. Epub 2021/01/11.
[13] Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19:477–489. Epub 2019/05/01.
[14] Thornberry NA, Bull HG, Calaycay JR, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. Epub 1992/04/30.
[15] Siu KL, Yuen KS, Castano-Rodriguez C, et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. Epub 2019/04/30.
[16] Kato K, Ikliptikawati DK, Kobayashi A, et al. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex. Biochem Biophys Res Commun. 2021;536:59–66. Epub 2020/12/29.
[17] Addetia A, Lieberman NAP, Phung Q, et al. SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98. mBio. 2021;12(2):e00065- Epub 2021/04/15.
[18] Xia H, Cao Z, Xie X, et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. Epub 2020/09/28.
[19] Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. Epub 2006/09/19.
[20] Miorin L, Kehrer T, Sanchez-Aparicio MT, et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci U S A. 2020;117:28344–28354. Epub 2020/10/25.
[21] Yoo JS, Sasaki M, Cho SX, et al. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat Commun. 2021;12:6602. Epub 2021/11/17.
[22] Lee JG, Huang W, Lee H, et al. Characterization of SARS-CoV-2 proteins reveals Orf6 pathogenicity,: subcellular localization, host interactions and attenuation by Selinexor. Cell Biosci. 2021;11:58. Epub 2021/03/27.
[23] Martin-Sancho L, Lewinski MK, Pache L, et al. Functional landscape of SARS-CoV-2 cellular restriction. Mol Cell. 2021;81:2656–2668. Epub 2021/05/01.
[24] Cao Z, Xia H, Rajsbaum R, et al. Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. Cell Mol Immunol. 2021;18:746–748. Epub 2021/01/22.
[25] Schaecher SR, Mackenzie JM, Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J Virol. 2007;81:718–731. Epub 2006/11/03.
[26] Yang R, Zhao Q, Rao J, et al. SARS-CoV-2 accessory protein ORF7b mediates tumor necrosis factor-alpha-induced apoptosis in cells. Front Microbiol. 2021;12:654709. Epub 2021/09/07.
[27] Stukalov A, Girault V, Grass V, et al.Multilevel proteomics reveals host perturbations by SARSCoV-2 and SARS-CoV. Nature. 2021;594:246–252. Epub 2021/04/13.
[28] Chen S, Zheng X, Zhu J, et al. Extended ORF8 gene region is valuable in the epidemiological investigation of severe acute respiratory syndrome-similar coronavirus. J Infect Dis. 2020;222:223–233. Epub 2020/05/21.
[29] Matsuoka K, Imahashi N, Ohno M, et al. SARS-CoV-2 accessory protein ORF8 is secreted extracellularly as a glycoprotein homodimer. J Biol Chem. 2022;298(3):101724, Epub 2022/02/15.
[30] Wu X, Xia T, Shin WJ, et al. Viral mimicry of interleukin-17A by SARS-CoV-2 ORF8. mBio. 2022;13(2): e0040222. Epub 2022/03/29.
[31] Lin X, Fu B, Yin S, et al. ORF8 contributes to cytokine stormduring SARS-CoV-2 infection by activating IL-17 pathway. iScience. 2021;24:102293. Epub 2021/03/17.
[32] Chou JM, Tsai JL, Hung JN, et al. The ORF8 protein of SARS-CoV-2 modulates the spike protein and its implications in viral transmission. Front Microbiol. 2022;13:883597. Epub 2022/06/07.
[33] Zhang Y, Chen Y, Li Y, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-iota. Proc Natl Acad Sci U S A. 2021;118(23):e2024202118. Epub 2021/05/23.
[34] Kee J, Thudium S, Renner DM, et al. SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature. 2022;610:381–388. Epub 2022/10/06.
[35] Leung K, Shum MH, Leung GM, et al. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26(1). Epub 2021/01/09.
[36] Nagy A, Pongor S, Gyorffy B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int J Antimicrob Agents. 2021;57:106272. Epub 2020/12/22.
[37] Xu K, Zheng BJ, Zeng R, et al. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology. 2009;388:279–285. Epub 2009/04/28.
[38] NeupertW, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. Epub 2007/02/01.
[39] Drozdetskiy A, Cole C, Procter J, et al. Pred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. Epub 2015/04/18.
[40] Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure predictionwithAlphaFold. Nature. 2021;596:583–589. Epub 2021/07/16.
[41] Mirdita M, Schütze K, Moriwaki Y, et al. ColabFold – making protein folding accessible to all. bioRxiv. 2022:2021.08.15.456425.
[42] Teufel F, Almagro Armenteros JJ, Johansen AR, et al. Signalp 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol. 2022;40(7):1023–1025. Epub 2022/01/05.
[43] Lin X, Fu B, Xiong Y, et al. Unconventional secretion of unglycosylated ORF8 is critical for the cytokine storm during SARS-CoV-2 infection. bioRxiv. 2021.
[44] Meier C, Aricescu AR, Assenberg R, et al. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. Epub 2006/07/18.
[45] Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. Epub 2007/08/08.
[46] Johnson HM, Torres BA, Soos JM. Superantigens: structure and relevance to human disease. Proc Soc Exp Biol Med. 1996;212:99–109. Epub 1996/06/01.
[47] Tomasello G, Armenia I,Molla G. The protein imager: a full-featured online molecular viewer interface with server-side HQ-rendering capabilities. Bioinformatics. 2020;36:2909–2911. Epub 2020/01/14.
The genome of the novel SARS-CoV-2 codes for an ORF1a/ ORF1ab (open reading frame) polyprotein containing sixteen non-structural proteins (NSP) and four structural proteins. The genome also has multiple ORFs coding for accessory proteins through a frame shift. These accessory proteins are not necessary for viral replication but might play a key role in pathogenesis of SARS-CoV-2. One such protein is the accessory protein 7a, which is predicted to contribute to Covid-19 by inducing the apoptotic processes in human host cells1.
Structure
SARS-CoV-2 is a very young virus and the structure and function of the accessory protein 7a has not yet been solved. However, 7a of SARS-CoV-2 shows 85% sequence identity and 95.2% sequence similarity with another protein in SARS-CoV2. It is therefore conceivable that both accessory proteins have a similar structure and function. The sequence analysis of SARS-CoV predicts that ORF7a codes for a type I transmembrane protein with 122 amino acids, including a signal peptide at the N‑terminus and a retrieval signal at the C-terminus3. The N-terminal ectodomain of ORF7a consists of seven β-strands compactly arranged in an immuno-globulin-like β-sandwich fold (Fig 1). These seven β-strands are ordered in two β-sheets containing four β-strands (A; G; F; C) in the first sheet and three (B; E; D) in the second one (see Fig 1: left)4.
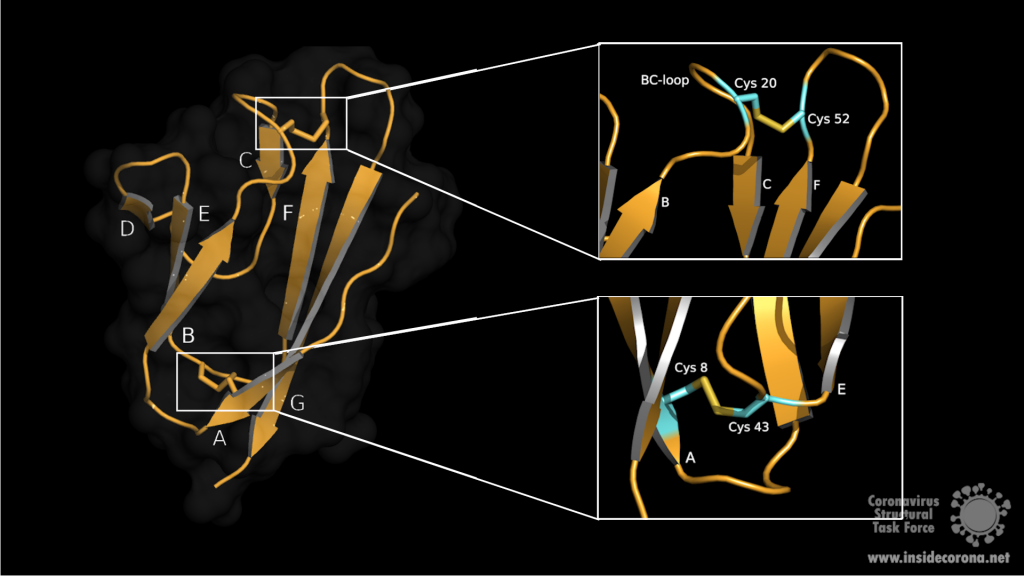
Both sheets are amphipathic, with the hydrophobic side facing inwards closely packed against each other. The top of the ectodomain is defined by the BC, DE and FG loops and the bottom by the AB, CD and EF loops. The β-sandwich structure is stabilized by two disulphide bonds linking the sheets at opposite edges. At the bottom of the structure, a disulphide bridge connects Cys8 on strand A with Cys43 at the end of strand E. At the top, Cys20 of the BC loop is linked to Cys52 at the end of strand F (see Fig 1: right). Additionaly, on top of the BED sheet, the DE loop protrudes from the structure and forms a groove together with β-strands C and D. In the centre is Glu18 which contributes to the negatively charged bottom of the mainly hydrophobic groove. This grove may be a potential site for ligand interaction due to its central negative electrostatic potential4.
Function
In cell culture, the polypeptide 7a of SARS-CoV seems to have diverse biological functions5. It is possible that 7a plays a key role in cell cycle control. In HEK 293 cells, an overexpression of 7a led to inhibition of cell growth and induction of the G0/G1 phase cell cycle arrest. This arrest may favour coronavirus replication and exacerbate virus-induced pathogenicity. 7a is also predicted to induce apoptosis in human kidney epithelial cells by interaction with a protein called B-cell lymphoma-extra large (Bcl-XL). Bcl-XL belongs to a group of pro-survival proteins, the B-cell lymphoma-2 (Bcl-2)- family, which prevent apoptosis in epithelial cells. The Interaction between 7a and the C-terminal transmembrane domain of Bcl-XL may interfere with this pro-survival function, leading to apoptosis via the caspase-dependant pathway6,7. In addition to this, SARS 7a interacts with a Ap4A-hydrolase involved in cell proliferation, DNA-replication, apoptosis and RNA-processing. This interaction leads to downregulation of its hydrolase-activity and an increased production of AP4A (diadenosine tetraphosphate) which may also induce apoptosis5. Such a host cell specific modulation of apoptosis could enable the virus to evade the immune response or to spread to other target organs.
Another predicted function of ORF7a is the inhibition of the bone marrow matrix antigen 2 (BST-2) that might restrict virus release by physically tethering the budding enveloped virion to the plasma membrane. ORF7a antagonizes this function by binding of the extracellular domain of BST-2 preventing its glycosylation. Thus, an inhibitor preventing ORF7a-BST-2 interaction can be speculated as potential drug target8.
Taken together, ORF7a is a virulence factor that contributes in different ways to the pathogenicity of SARS-CoV-2. Therefore, targeted drug development against ORF7a could be a critical factor to reduce viral spread or attenuate severe disease progression.
PDB Structures Available
6W37: X-ray structure of the SARS-CoV-2 ORF7a encoded accessory protein.
1xak: SARS-CoV ORF7a accessory protein, a unique type I transmembrane protein of unknown function. Has a short cytoplasmic tail and a transmembrane domain. Consists of one chain (chain A), that forms a compact seven-stranded beta sandwich.
1y04: SARS Coronavirus ORF 7a coded X4 protein, also known as 7a, U122 or X4. Type-I transmembrane protein with immunoglobulin like beta-sandwich fold. Potential functions of X4 in virus replication and pathogenesis are discussed.
References
- 1.Michel CJ, Mayer C, Poch O, Thompson JD. Characterization of accessory genes in coronavirus genomes. Virol J. Published online August 27, 2020. doi:10.1186/s12985-020-01402-1
- 2.Francis K. Y. The Proteins of Severe Acute Respiratory Syndrome Coronavirus‑2 (SARS CoV‑2 or n‑COV19), the Cause of COVID‑19. The Protein Journal (2020). 2020;(39):198-216. doi:10.1007/s10930-020-09901-4
- 3.Fielding BC, Tan Y-J, Shuo S, et al. Characterization of a Unique Group-Specific Protein (U122) of the Severe Acute Respiratory Syndrome Coronavirus. JVI. Published online July 15, 2004:7311-7318. doi:10.1128/jvi.78.14.7311-7318.2004
- 4.Hänel K, Stangler T, Stoldt M, Willbold D. Solution structure of the X4 protein coded by the SARS related coronavirus reveals an immunoglobulin like fold and suggests a binding activity to integrin I domains. J Biomed Sci. Published online November 23, 2005:281-293. doi:10.1007/s11373-005-9043-9
- 5.Vasilenko N, Moshynskyy I, Zakhartchouk A. SARS coronavirus protein 7a interacts with human Ap4A-hydrolase. Virology Journal. Published online 2010:31. doi:10.1186/1743-422x-7-31
- 6.Tan Y-J, Fielding BC, Goh P-Y, et al. Overexpression of 7a, a Protein Specifically Encoded by the Severe Acute Respiratory Syndrome Coronavirus, Induces Apoptosis via a Caspase-Dependent Pathway. JVI. Published online December 15, 2004:14043-14047. doi:10.1128/jvi.78.24.14043-14047.2004
- 7.Tan Y-X, Tan THP, Lee MJ-R, et al. Induction of Apoptosis by the Severe Acute Respiratory Syndrome Coronavirus 7a Protein Is Dependent on Its Interaction with the Bcl-XL Protein. JVI. Published online April 11, 2007:6346-6355. doi:10.1128/jvi.00090-07
- 8.Taylor JK, Coleman CM, Postel S, et al. Severe Acute Respiratory Syndrome Coronavirus ORF7a Inhibits Bone Marrow Stromal Antigen 2 Virion Tethering through a Novel Mechanism of Glycosylation Interference. García-Sastre A, ed. J Virol. Published online September 16, 2015:11820-11833. doi:10.1128/jvi.02274-15
