Für diesen Beitrag exisitiert leider keine deutsche Übersetzung.
Introduction
This protein is known under many different names such as non-structural protein NSP1, leader protein, host translation inhibitor and host shutoff factor. Some of these names already tell us about the function and importance of this relatively small protein. It is found in all betacoronaviruses1 and, even though it only contains 180 amino acids2, it is indispensable for the viral life cycle and the pathogenicity of SARS-CoV-2.
It plays an important role when it comes to the point where the virus needs its own genetic information in form of a string of codons. Its mRNA is translated into the corresponding amino acids that make up the viral proteins. Translation occurs either shortly after the virus entered the host cell (see life cycle) or after the viral mRNA has been replicated (as described here).
For this process, the virus does not have its own proteins; instead, it just uses the already existing translation machinery of the host cell: the ribosomes.
As ribosomes are responsible for synthesizing proteins by translating the information on the host’s mRNA into a string of amino acids, they are an important part of human cells. They consist of ribosomal RNA (rRNA) and ribosomal proteins, which form a larger (60S) and a smaller (40S) subunit3.
Here, the NSP1 comes into play. It helps the virus hijack ribosomes and use them for the replication of its own mRNA, while the host cells translation is supressed/inhibited/shut off4.
To understand how the NSP1 is involved in all this, we will first have take a closer look at the structure of the protein.
Structural features & interaction with ribosomes
Even though the full-length structure of NSP1 is unknown so far, we know what the two individual domains (connected via a linker that is 20 amino acids long) of the SARS-CoV-2 NSP1 look like and can even say a lot about its interaction with human ribosomes.
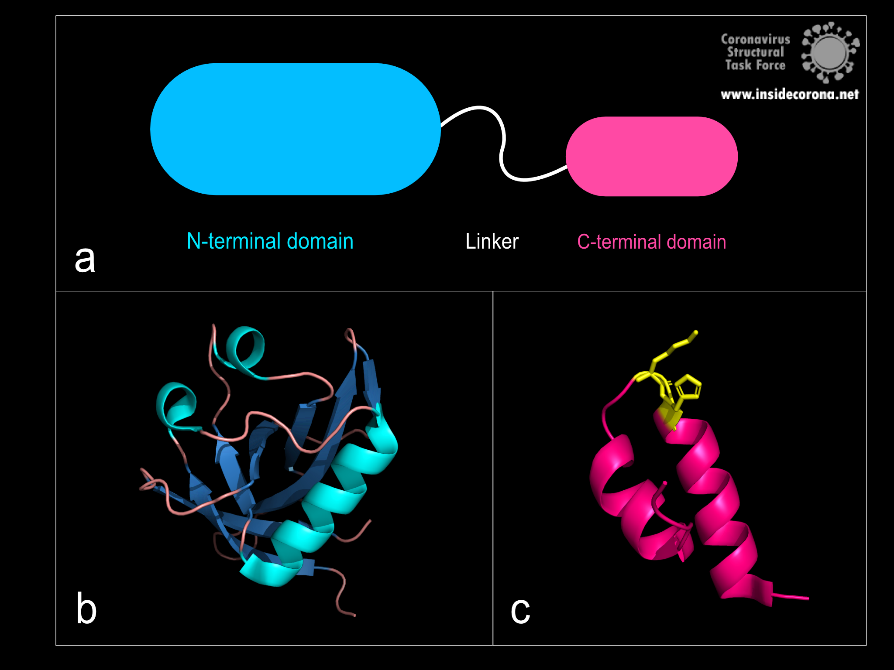
Figure 1: a: Schematic structure of NSP1. b: N-terminal domain (PDB: 7K7P)., c: C-terminal domain. KH motif (amino acids K164 and H165) in yellow (PDB: 6ZLW).
The first domain is the globular N-terminal domain (amino acids 1–128), which takes up most of the protein. It consists of a β-barrel of seven β-strands, two 310 helices and one α-helix5, as can be seen in Figure 1b.
The probably more interesting domain, due to the crucial role it plays for interaction with human ribosome, is the C-terminal domain comprising three moieties (Figure 1c). It consists of the two α-helices, α1 and α2, and a loop connecting them4. The shape of this C-terminal domain and its surface charge matches the mRNA entry channel of the ribosome perfectly and therefore covers the whole usual mRNA path4. In Figure 2, the small 40S ribosomal subunit (green) in a complex with the C-terminal domain of NSP1 (pink) is shown.
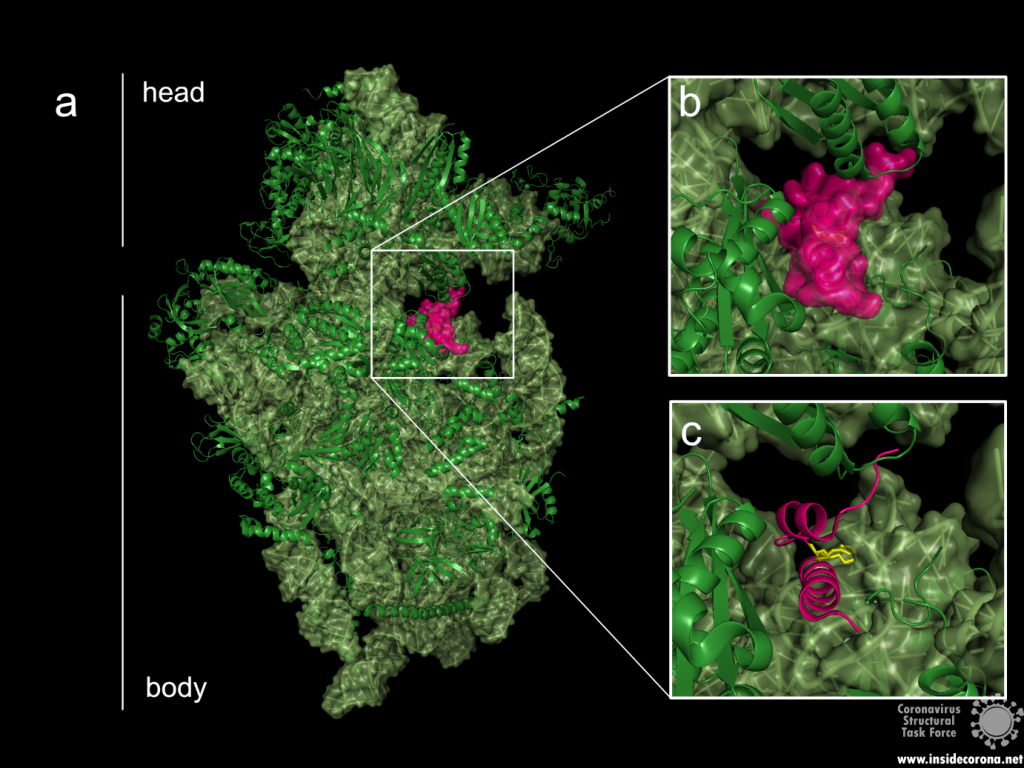
Figure 2: a: Ribosomal 40S subunit in complex with the NSP1 C-terminal domain (PDB: 6ZLW). The C-terminal domain is bound to the mRNA channel between the “head” and “body” of the 40S. b & c: NSP1 C-terminal domain shown with and without surface.
While the C-terminal domain is bound to the mRNA entry channel of the host cell’s 40S ribosomal subunit, the N-terminal domain can move around it within a 60 Å radius, connected by the 20 amino acid long flexible linker6.
All these interactions lead to an inhibition of the translation of the hosts mRNA—but how does the viral mRNA get translated, if the NSP1 is bound to the ribosome’s mRNA entry channel?
Viral translation
The virus needs a mechanism to circumvent its own translational blockage to maintain the capability for translation of the viral mRNA. It is not yet completely clear how this is accomplished, but different suggestions exist.
The first theory involves the N-terminal domain of NSP1 and the 5’ untranslated region (5’UTR) of the viral mRNA7.
In most coronaviruses, the 5’UTR part of the viral mRNA is conserved with a complex secondary structure6. Some scientists7 suggest that it might interact with the N-terminal domain, making the interaction between NSP1 and the ribosome sterically impossible and therefore lifting the blockage. This was also based on their study indicating that the C-terminal domain alone can suppress the host’s protein synthesis, but the N-terminal domain is needed to bypass the translation inhibition. Also, extending the linker between the two domains artificially by additional amino acids could be shown to reduce the viral mRNA translation7.
The second theory suggests that the translational blockage induced by the viral NSP1 is not lifted. In this mechanism, most ribosomes would be blocked by the NSP1s, but those left unblocked could still synthesize proteins. Here the viral 5’UTRs would make the mRNA of the virus more favourable than the host’s mRNA. This would lead the ribosomes into translating the viral mRNA with a higher efficiency than the cellular mRNA6.
Effect on the cells and immune system interference
Translation inhibition of the cellular mRNA by NSP1 results directly in another interesting and significant effect on the human cell. Besides the negative effects on normal cell functions, the translation of proteins involved in innate immune response is also inhibited. This includes interferons (proteins involved in antiviral activity8) like Interleukin-8, IFN-β, IFN-γ1 and anti-viral factors that are stimulated by interferons, leading to a downregulation of the cell’s defence system4,9.
Earlier studies on SARS-CoV-1 also showed that NSP1 is further inducing cleavage of the host’s mRNA, probably by using one of the host’s proteins. This again does not apply to its own viral mRNA10, making the impact on the host cell even greater.
Taken together, this protein is a major pathogenicity factor of SARS-CoV-2 and might therefore be an interesting drug target1.
Available structures
As of this writing, 16 structures of the SARS-CoV-2 NSP1 are available, of which two display the N-terminal domain. The other structures show the C-terminal domain in complex with a ribosome, ribosomal subunit or preinitiation ribosome. As there is no full-length structure solved so far, only predictions on the whole protein were made, for example given by Clark et al.5.
Available structures of the N-terminal: 7k7p, 7k3n.
Available structures of the C-terminal: 7k5i, 6zoj, 6zok, 6zm7, 6zlw, 6zmi, 6zp4, 6zon, 7jqb, 6zme, 6zmt, 6zn5, 6zmo, 7jpc.
References
- de Lima Menezes, G. & da Silva, R. A. Identification of potential drugs against SARS-CoV-2 non-structural protein 1 (nsp1). Journal of Biomolecular Structure and Dynamics 1–11 (2020) doi:10.1080/07391102.2020.1792992.
- Yoshimoto, F. K. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. 19.
- Khatter, H., Myasnikov, A. G., Natchiar, S. K. & Klaholz, B. P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015).
- Thoms, M. et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. 8 (2020).
- Clark, L. K., Green, T. J. & Petit, C. M. Structure of Nonstructural Protein 1 from SARS-CoV-2. Journal of Virology 95, 12 (2021).
- Schubert, K. et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27, 959–966 (2020).
- Shi, M. et al. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. http://biorxiv.org/lookup/doi/10.1101/2020.09.18.302901 (2020) doi:10.1101/2020.09.18.302901.
- De Andrea M. et al. The interferon system: an overview. Eur J Paediatr Neurol (2002) doi:10.1053/ejpn.2002.0573.
- Vann, K. R. Inhibition of translation and immune responses by the virulence factor Nsp1 of SARS-CoV-2. 4.
- Huang, C. et al. SARS Coronavirus nsp1 Protein Induces Template-Dependent Endonucleolytic Cleavage of mRNAs: Viral mRNAs Are Resistant to nsp1-Induced RNA Cleavage. PLoS Pathog 7, e1002433 (2011).
