Für diesen Beitrag exisitiert leider keine deutsche Übersetzung.
This article has been written by Cameron Fyfe and Lea von Soosten.
In the previous two articles we spoke of proteins involved in RNA synthesis and proteins involved in removing errors during that process. There are also proteins produced by SARS-CoV-2 that can mimic functions of the host cell to avoid its defense mechanisms.
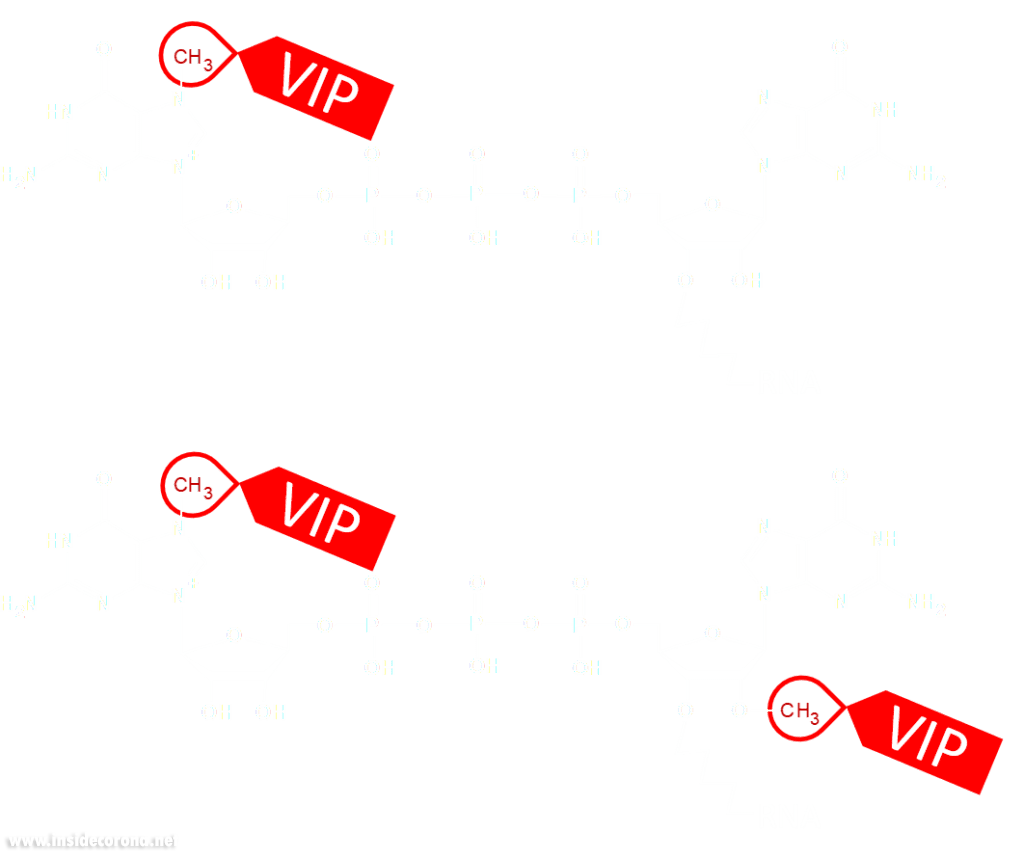
Eukaryotic cells have evolved to have various immune responses to fight infection or invasion from pathogens. One of these is to recognize and chop up any RNA that is from other organisms using enzymes called exoribonucleases. In order to differentiate "friendly" RNA from "foe" RNA is to give the cell's own RNA a VIP badge so that only unfriendly RNA will be shredded. These "VIP badges" are made of a 5’ to 5’ triphosphate linkage with two methylation modifications (see Fig. 1). In order to evade exoribonucleases, the virus SARS-CoV-2 has a way of 5’ to 5’ capping as well as adding its own methyl group VIP badges to protect its RNA from the defense mechanisms of invaded cells. Two Very Important Proteins, nsp14 and nsp16, have this methyltransferase activity using an S-Adenosyl methionine (SAM) as cofactor.
What are SAM methyltransferases?
Methyltransferase enzymes are a large superfamily of proteins that perform the chemical addition of a methyl group (a carbon with three hydrogens) to a variety of substrates. These substrates include small molecules, other proteins, DNA, and RNA 2,3. This superfamily of proteins often uses a small molecule, S-Adenosyl methionine (SAM), to transfer a methyl group to its target substrate (Figure 2). During this process, the methyl group bound to the charged sulfur is brought in proximity to the target atom of the substrate, transferring the methyl group (Figure 2), resulting in the methylated product and the byproduct S-Adenosyl homocysteine (SAH).
Methyltransferases of SARS-CoV-2
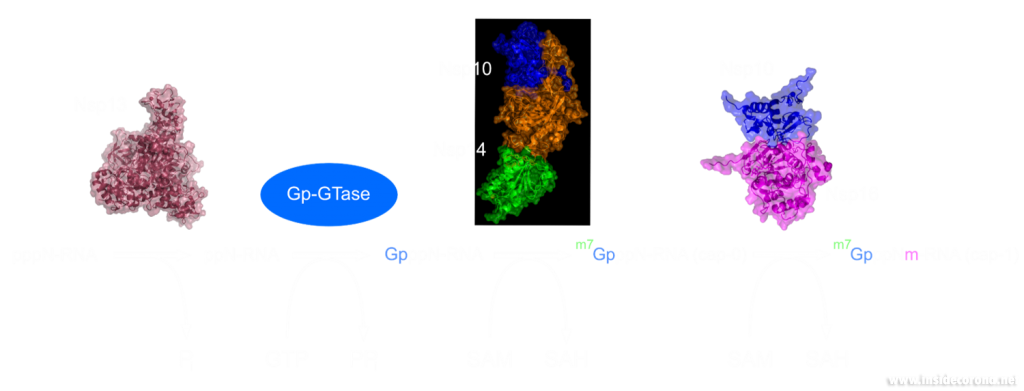
In a previous article we spoke of the exoribonuclease (ExoN) proofreading activity of Nsp14 (not to be confused with the host cell's own exoribonucleases that are part of the immune system, see above). After the 5’ to 5’ guanine triphosphate addition has been performed on the mRNA the guanine-N7-methyltransferase activity of Nsp14 comes into play producing the first Cap0 structure with a VIP tag (Figure 1, 3). Only after this methylation has been performed can Nsp16 have action and perform the second 2’O-methylation to produce the Cap1 structure (Figure 1, 3).
Not only do both of these proteins perform VIP methylations of mRNA, but they also both bind another non-structural protein, Nsp10. The binding of Nsp10 has been shown to increase activity in both Nsp14 ExoN activity and Nsp16 methyltransferase activity4. Independently, Nsp10 has also been shown to have the ability to bind both single and double stranded DNA and RNA5.
Structures of nsp14 and nsp16
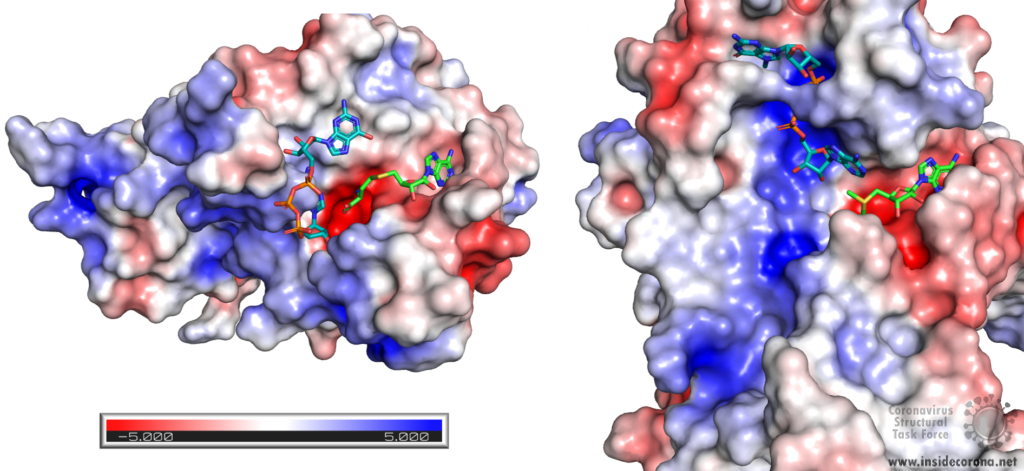
Nsp14 consists of two domains, each carrying out one specific task: the first is responsible for the ExoN activity, whilst the second executes the first methylation of the Guanosine-N7 of the RNA end cap. The two domains are connected by a flexible region that acts like a hinge, allowing movement between the domains. The second domain has an unusual and unique structure which does not follow the typical Rossmann fold seen in other SAM methyltransferases. The methyltransferase active site has a negatively charged binding pocket that holds SAM (SAH in Figure 4. A) in close proximity to the Guanosine-P3-adenosine-5',5'-triphosphate (GpppA) substrate (Figure 4A). The binding pocket holding the GpppA has a positive charge and the surface charge of the region below is also positively charged (Figure 4A). The distance between the N7 of the 5’ Guanosine and the sulfur that transfers the methyl group is 4.4 Å5,6. This close proximity of cofactor and substrate facilitates the methylation.
Similar to Nsp14, Nsp16 has a negatively charged binding pocket to position SAM in close proximity to the m7GpppA substrate (Figure 4. B). The m7GpppA binding site has a positive charge. The space nearby the 3’ end of the m7GpppA also has an overall positive charge and would be expected to bind the extension of the full length RNA (Figure 4. B)4. The distance between the methyl group and the sulfur of SAM and the 2’O of the m7GpppA substrate is 3.1Å and 4.9Å, respectively.
Structure of nsp10 and its function
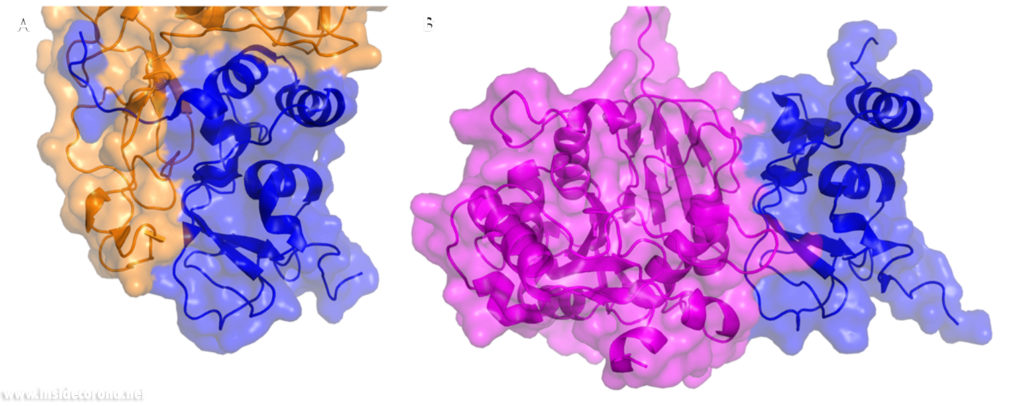
In a previous article where we spoke about the exoribonuclease (ExoN) activity of the first domain of nsp14, we highlighted the interaction between nsp14 and nsp10 (Figure 5A). This is quite significant, as the activity of ExoN increases 30-fold when nsp10 and nsp14 are bound. Nsp10 also functions as a co-factor for nsp16, stabilizing the SAM-binding pocket7 and enhances its methyltransferase enzymatic activity significantly4 (Figure 5B). For SARS-CoV, and similarly for MERS-CoV, the affinity for m7GpppA-RNA and m7GpppA cap analogue of nsp16 was found to be low until binding to nsp10, which enhanced the affinity for binding to RNA8,9. With a reduced activity in Nsp16 in the absence of Nsp10 and a huge decrease in activity of the exonuclease domain of Nsp14, interfering with these interactions could result in decreased viability of COVID-19.
Methyltransferases Nsp14 and Nsp16 as drugs targets
As both Nsp14 and Nsp16 use the cofactor SAM and have affinity for the endcap of RNA, these two binding sites could be worthwhile targets for drug development in the fight against SARS-CoV-2. Without the VIP status provided by the methylation of RNA the host immune system could defend against the viral RNA. It might be possible to block these binding pockets by letting the protein bind to something that is similar to SAM, which cannot function as a methyl donor. An additional challenge is that the inhibitor has to be very specific to Nsp14 or Nsp16, so as not to affect similar human proteins in a negative way.
Sinefungin is a 5’-aminoalkyl analog of SAH and SAM, which can do exactly that: it has the ability to inhibit all SAM methyltransferases (Figure 6). Sinefungin was first discovered in 1973 from Strepromyces griseolus and was described as having antifungal antibiotic properties10.
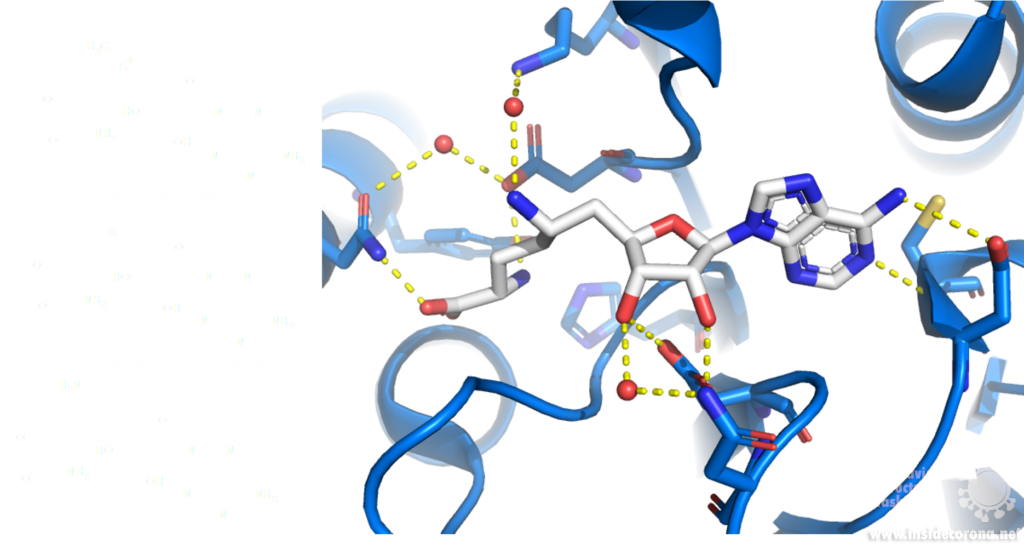
A major issue with targeting the SAM binding site of Nsps with compounds such as sinefungin (Figure 6) is that there are many proteins within humans that use SAM as a cofactor for normal function. This results in singefungin and other similar compounds having toxic effects on human cells. Synthetic chemists have already been able to synthesize analogs of sinefungin with improved affinities to specific SAM methyltransferases. Recently, specific inhibitors have been developed to target a nicotinamide SAM methyltransferase11. This inhibitor was developed to have affinity to both the cofactor binding site and the substrate binding site by combining the nicotinamide substrate with the SAM cofactor. Recent work has looked at how singefungin binds to the active site of Nsp16 in order to have a detailed understanding of its interaction to design more specific inhibitors that can target methyltransferases from SARS-CoV-24. Similar to the development of the nicotinamide SAM methyltransferase inhibitor, developing an inhibitor which binds to the substrate binding site as well as to the cofactor binding site could be effective. As Nsp14 and Nsp16 target different substrates, any inhibitors designed in this way would likely have specificity to only one of the two methyltransferases from SARS-CoV-2. Of the two, Nsp14 might be easier to target as it has a unique structure not similar to human SAM methyltransferases.
As both Nsp14 and Nsp16 interact with Nsp10 for normal function, interfering with this interaction could reduce activity of these enzymes. Further still, as the interface between Nsp10 with Nsp14 and Nsp16 has overlap the target is smaller for blocking binding of these proteins.
One way to look for possible drugs is repurposing those which are already approved for other diseases. Initial screen can be done in silico, by simulations of the interaction between the protein and the already existing and approved drug. However, such studies are highly dependent on the protein structures employed being correct, which is why we are evaluating all structures that are published for SARS-CoV and SARS-CoV-2.
Available structures
If you would like to look at the currently available structures for Nsp10, Nsp14, and Nsp16, they are available from our data base; we provide information on the quality of measurement data and models as well as improved structures.
All structures available for Nsp14 are bound to Nsp10 and are only available from SARS-CoV. The highest resolution structure of Nsp14 is PDB entry 5c8t at 3.2Å. It has a bound S-Adenosyl methionine ligand as well as zinc ions present. Alongside this, another structure of Nsp14 bound to S-Adenosyl homocysteine and a guanosine-triphosphate-adenosine ligand as well as zinc at 3.33Å resolution has been published (PDB: 5c8s). Additionally, two structures with zinc atoms but no ligands are available (PDB 5c8u 3.4Å at and 5nfy at 3.34Å). Both PDB entries 5c8t and 5nfy have been improved structures by our group.
Similar to Nsp14 all structures of Nsp16 are bound to Nsp10. There are currently 18 structures for Nsp16 bound to Nsp10 from SARS-CoV-2. The highest resolution structure is at 1.8Å and has SAM, Guanosine triphosphate and Adenosine bound as well as zinc atoms. The PDB:6wkq has Nsp16 bound to the methyltransferase inhibitor Sinefungin at 1.98Å resolution. Two further structures of note are 7jhe and 7jib that have various functional ligands. A further four structures are available from SARS-CoV.
Nsp10 alone: Currently there are two structures of Nsp10 from SARS-CoV-2, PDB 6zpe and 6zct, with the former having the highest resolution of 1.58 Å with bound zinc (PDB 6zpe). There are also three structures of Nsp10 from SARS-CoV available, PDB 2fyg, 2g9t, and 2ga6.
- 1.Ramanathan A, Robb GB, Chan S-H. mRNA capping: biological functions and applications. Nucleic Acids Res. Published online June 17, 2016:7511-7526. doi:10.1093/nar/gkw551
- 2.Boriack-Sjodin PA, Swinger KK. Protein Methyltransferases: A Distinct, Diverse, and Dynamic Family of Enzymes. Biochemistry. Published online December 22, 2015:1557-1569. doi:10.1021/acs.biochem.5b01129
- 3.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. Published online October 16, 2017:81-92. doi:10.1038/nrg.2017.80
- 4.Krafcikova P, Silhan J, Nencka R, Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun. Published online July 24, 2020. doi:10.1038/s41467-020-17495-9
- 5.Ferron F, Subissi L, Silveira De Morais AT, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci USA. Published online December 26, 2017:E162-E171. doi:10.1073/pnas.1718806115
- 6.Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc Natl Acad Sci USA. Published online July 9, 2015:9436-9441. doi:10.1073/pnas.1508686112
- 7.Rosas-Lemus M, Minasov G, Shuvalova L, et al. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. Published online April 20, 2020. doi:10.1101/2020.04.17.047498
- 8.Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells. Published online May 20, 2020:1267. doi:10.3390/cells9051267
- 9.Chen Y, Su C, Ke M, et al. Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex. Kuhn RJ, ed. PLoS Pathog. Published online October 13, 2011:e1002294. doi:10.1371/journal.ppat.1002294
- 10.Robert L. H, Marvin M. H. A9145, A NEW ADENINE-CONTAINING ANTIFUNGAL ANTIBIOTIC. J Antibiot. 1973;26(8):463-465. doi:10.7164/antibiotics.26.463
- 11.Policarpo RL, Decultot L, May E, et al. High-Affinity Alkynyl Bisubstrate Inhibitors of Nicotinamide N-Methyltransferase (NNMT). J Med Chem. Published online October 7, 2019:9837-9873. doi:10.1021/acs.jmedchem.9b01238
