Für diesen Beitrag exisitiert leider keine deutsche Übersetzung.
This article has been written for the journal Crystallographic Reviews and has been published on 11 June 2022 online. For additional information, data and tables, please see the full publication: https://doi.org/10.1080/0889311X.2022.2065270.
Abstract
The SARS-CoV-2’s endoribonuclease (NendoU) nsp15, is an Mn2+ dependent endoribonuclease specific to uridylate that SARS-CoV-2 uses to avoid the innate immune response by managing the stray RNA generated during replication. As of the writing of this review 20 structures of SARS-CoV-2 nsp15 have been deposited into the PDB, largely solved using X-ray crystallography and some through Cryo-EM. These structures show that a nsp15 monomer consist of three conserved domains, the N-terminal oligomerization domain, the middle domain, and the catalytic NendoU domain. Enzymatically active nsp15 forms a hexamer through a dimer of trimers (point group 32), whose assembly is facilitated by the oligomerization domain. This review summarises the structural and functional information gained from SARS-CoV-2, SARs-CoV and MERS-CoV nsp15 structures, compiles the current structure-based drug design efforts, and complementary knowledge with a view to provide a clear starting point for downstream structure users interested in studying nsp15 as a novel drug target to treat COVID-19.
Introduction
SARS-CoV-2 is a nidovirus with a non-segmented positive-sense RNA genome, meaning the RNA genome is read from 5′ to 3′ and can be directly translated into viral proteins; it’s effectively messenger RNA. The RNA genome of SARS-CoV-2 is one of the largest RNA genomes among RNA viruses [1], comprised of a replicase gene which encodes non-structural proteins (nsps), structural proteins, and accessory proteins. The genome can produce two different polyprotein chains through a ribosomal frameshift [2] (ORF1a and ORF1b). Once translated, these polyproteins are cleaved by one of the two encoded proteases (3C-like protease (nsp5) or papain-like-protease (nsp3)) to yield between 15 and 16 non-structural proteins, which assemble into a large membrane-bound replicase complex (RTC).
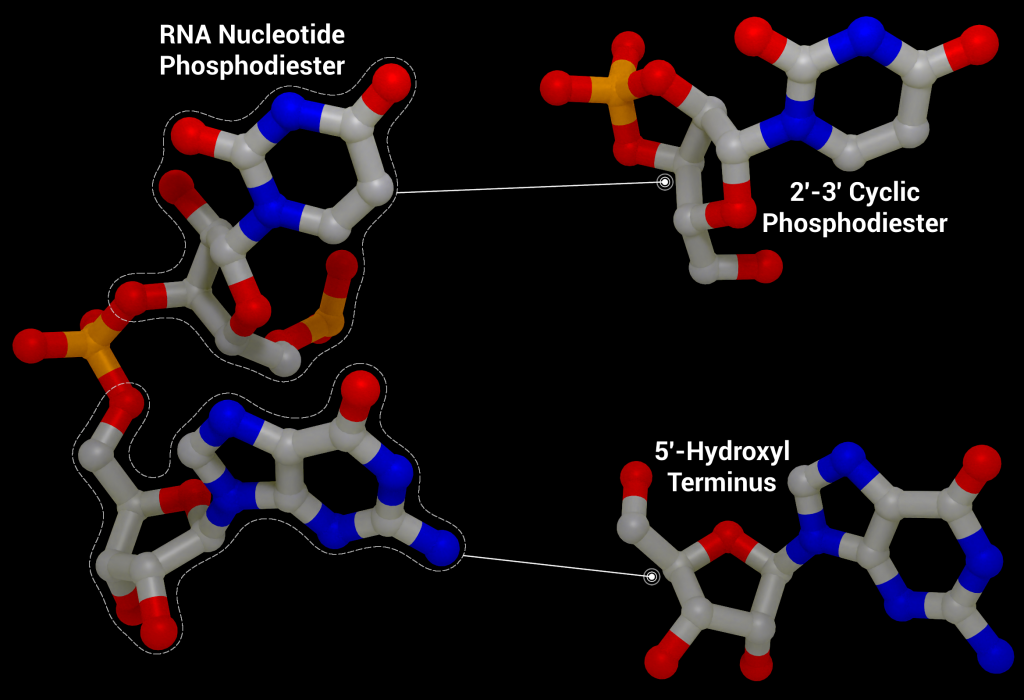
One of these non-structural proteins is nsp15, a 346 amino acid nidoviral RNA uridylate‐specific and Mn2+-dependent [3] endoribonuclease (NendoU). Its gene is found towards the end of the non-structural proteins in the SARS-CoV-2 genome on ORF1b (bases 6453 to 6798[4]). Nsp15 preferentially cleaves the 3′ end of uridine, producing a 2′‐3′ cyclic phosphodiester and 5′‐hydroxyl terminus [5] (Figure 1). Nsp15 is conserved across coronavirus family members [6], to the point where it has been proposed as a universal genetic marker to distinguish nidoviruses [7] from all other RNA virus families. Although highly conserved (88% sequence identity with SARS-CoV-2, 50% with MERS, and 43% with HCoV-229E), nsp15 has been found to be non-essential for viral replication in Mouse Hepatitis Virus [6] (MHV), SARS-CoV, and HCoV-229E. Some nsp15 mutations completely abolished RNA synthesis; however, these mutations resulted in misfolded and insoluble nsp15 when expressed in E. coli[6]. As a result, the loss of RNA synthesis is thought to be a knock-on effect on neighbouring polyprotein components that are critical for replication, as opposed to a genuine effect on viral replication through lack of nsp15 [6]. Further evidence of nsp15’s non-essential role in viral replication comes from insect nidoviruses and invertebrate roniviruses, which completely lack EndoU activity [8,9].
Although not essential for viral replication, recent studies suggest nsp15 plays a role in repressing activation of the host innate immune response [11–13]. During viral replication, positive-sense RNA is translated to produce the viral replication complex, which replicates the positive-sense RNA to produce negative-sense RNA. The negative-sense RNA then acts as a template to produce new positive-sense genomic RNA and subgenomic RNA. Subgenomic RNA consists of smaller transcribed sections of RNA produced by initiating transcription in the middle of the template strand (internal initiation), falling off the template strand before reaching the 5’ stop codon (premature termination), or by jumping off the template strand and reinitiating transcription further down the template (discontinuous transcription). This process produces short and long double-stranded RNA intermediates with polyuridine tracts at the 5′ end which can be recognized by pattern recognition receptors in the host cell such as RIG-I-like receptors (RLRs), protein kinase R (PKR), oligoadenylate synthases (OASes), and melanoma differentiation-associated gene 5 (MDA5). These sensors promote an innate and antiviral immune response [11,14,15] by activating the type I and III interferon (IFN) response, which induces expression of interferon -stimulated genes through the signal transducer and activator of transcription proteins 1 and 2 (STAT1/2) signaling pathways. By cleaving the 5′-polyuridine tracts in negative-sense viral RNA, nsp15, along with nsp16 and nsp10, limit the accumulation of MDA5-dependent pathogen-associated molecular patterns to delay the host’s immune response [16]. Loss of nsp15 activity has been shown to activate the interferon response and reduce viral titers in piglets infected with nsp15-deficient porcine epidemic diarrhea coronavirus (PEDV) [17] and mice infected with nsp15-deficient Mouse Hepatitis Virus [11]. It has also been demonstrated that nsp15 plays a role in disrupting formation of autophagosomes, which are double-membraned vesicles containing cellular material to be degraded.
Structural overview
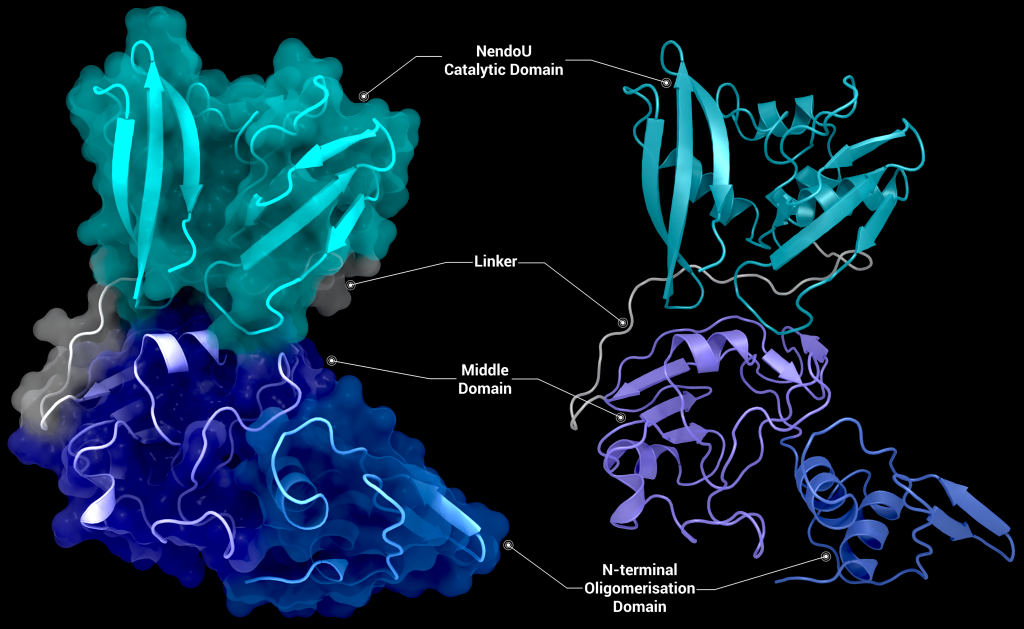
SARS-CoV-2 nsp15 consists of an N-terminal oligomerisation domain (Figure 2, blue), a middle domain (Figure 2, purple), and the catalytic C-terminal NendoU domain (Figure 2, teal). The Oligomerisation domain is formed from an anti-parallel β-sheet (β1-3) which wraps around helices α1 and α2. The middle domain consists of three β-hairpins (β5-6, β7-8, and β12-13), a mixed β-sheet (β4, β9, β10, β11, and β15), 2 α-helices (α3 and α4), and a right-handed 310 helix (η4). The catalytic NendoU domain contains two anti-parallel β-sheets (β16-18 and β19-21) which form a concave surface flanked by five α-helices (α6, α7, α8, α9, and α10). SARS-CoV-2 nsp15 shows high sequence identity with SARS-CoV nsp15 (88%) and lower sequence identity with MERS-CoV (51%), however the overall structural similarity is very high between the three viruses [1]. Three structures have been solved for SARS-CoV nsp15 (PDB entries 2H85 [18], 2OZK [19], and 2RHB [20]) one structure of MERS nsp15 (PDBID: 5YVD [21]), two structures from mouse hepatitis virus (2GTH and 2GTI [3]), and one structure from human coronavirus 229E (PDB entry 4S1T). As of writing this review 20 structures of SARS-CoV-2 nsp15 have been solved with a variety of bound ligands using X-ray crystallography and cryo-EM (Table 1) [1,22,23].
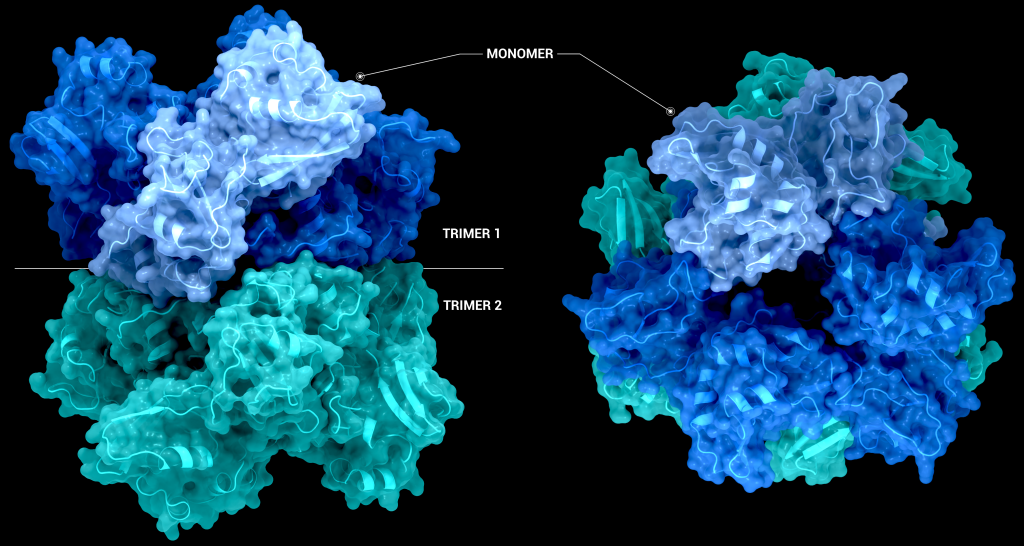
The biological assembly of nsp15 is a double-ringed hexamer made up of a dimer of trimers (point group 32, Figure 3). The trimeric form retains some ribonuclease activity, but the monomer presents with only residual cleavage [24] The hexamer is stabilised by an N-terminal oligomerisation domain present in each monomer. A crystal structure from SARS-CoV with a 28 amino acid N-terminal truncation (PDB entry 2H85) presented with a misfolded endoU active site, suggesting oligomerisation may act as an allosteric activation switch [19]. The six monomers come together to form the active enzyme with a 100 Å long negatively charged channel 10 to 15 Å wide open to solvent at the top, bottom, and on three separate side openings in the middle of the hexamer. Formation of the hexamer is essential for enzymatic activity, making the oligomerisation interfaces a potential target for structure-based drug design.
The active site of nsp15 is an electropositive pocket which lies at the interface between each monomer’s NendoU domain. The active site is highly conserved between SARS-CoV-2, SARS-CoV, and MERS proteins. Six key residues (His235, His250, Lys290, Thr341, Tyr343, and Ser294) are arranged in a shallow groove in the N-terminal NendoU domain [1]. His235, His250, and Lys290 are proposed to act as a catalytic triad, using a similar mechanism to that observed in RNase A [23]. However, RNase A is metal-independent, while SARS-CoV-2 nsp15 is Mn2+ dependent, so the mechanism is not an exact match. Mutation of either histidine in the catalytic triad to alanine eliminates RNA cleavage activity in nsp15 but has no effect on the formation of stable hexamers, showing they are not a factor in nsp15 oligomerisation [22]. This is unsurprising, as the N-terminal oligomerisation domain is the key player in the formation of the hexamer, but formation of the hexamer clearly plays an allosteric role in the formation of the active site, as activity in the monomer is significantly reduced.
Uracil specificity is proposed to be governed by Ser294 [20], with the main chain nitrogen of Ser294 predicted to interact with the carbonyl O2 oxygen of uracil and the hydroxyl group of Ser294 binding to uracil N3. However, mutation studies on homologs have shown that a Ser294Ala mutation significantly decreased activity without completely abolishing it[18] and negates uridine specificity. Tyr343 is likely important in governing uracil specificity, as shown by van der Waals stacking between the ribose sugar or Uridine and Tyr343 in cryo-EM structures [20,21]. Mutation of Tyr343 equivalent residues in SARS-CoV and MERS to alanine caused near complete loss of nuclease activity [20,21], suggesting a key role in enzymatic activity.
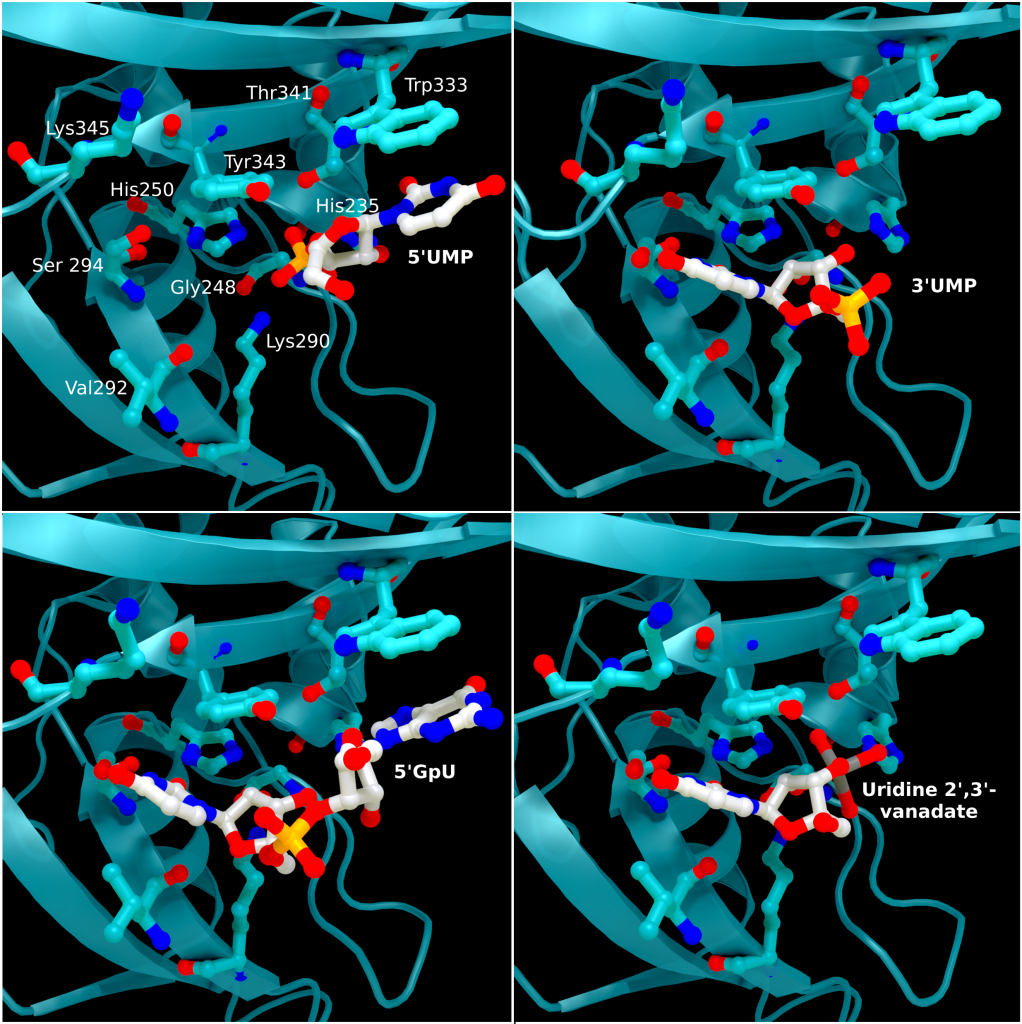
The structure of SARS-CoV-2 nsp15 has been solved in the presence of various catalytic intermediates, including 5′UMP (PDB entry: 6WLC), 3′UMP (PDB entry: 6X4I), 5′GpU (PDB entry: 6X1B), and uridine 2′,3′-vanadate (PDB entry: 7K1L). All intermediates bound to the C-terminal catalytic domain, interacting with the seven conserved active site residues (His235, His250, Lys290, Trp333, Thr341, Tyr343, Ser294, Gly248, Lys345, and Val292) and the structures showed no significant conformational deviations from each other (Cα RMSD = 0.29 Å). The uracil moiety of 5′UMP, guanylyl(3’-5’)uridine (GpU), and uridine 2′,3′-vanadate are all bound by Ser294 and Leu346 (Figure 4 Top left, bottom left, and bottom right, respectively), reinforcing the idea of uracil recognition being mediated by these residues. The combination of these structures confirms the predicted parallels between the reaction mechanism of SARS-CoV-2 nsp15 and RNAse A. The 5′UMP, 5′GpU, and uridine 2′-,3′-vanadate bound structures support the previously proposed hypothesis about uracil and purine base discrimination with Ser294 playing a key role [23]. Contrary to this finding, the 3′UMP bound structure shows the uracil base forming a stacking interaction with Trp333 (Figure 4, top right), the guanine binding site identified in the 5′GpU complex, suggesting nsp15’s active site can accommodate both purine and pyrimidine bases. However, the Trp333 interacting base is likely less relevant when binding larger RNA molecules as it provides a potential stacking interaction for bases without selectivity [23].
Comparison of these ligand-bound structures with RNase A catalytic sites suggests nsp15 acts through a similar reaction mechanism [23]. Based on these findings a two-step mechanism has been proposed starting with a transphosphorylation reaction whereby His250 acts as a base and deprotonates 2′OH of the RNA ribose, with Lys290 stabilising the negative charge that builds up during the transition state. His235 then acts as a general acid donating a proton for the departing 5′OH group. This is followed by a hydrolysis step where the roles of His250 and His235 are reversed, with His235 deprotonating a water molecule and His250 acing as a proton donor for the 5′OH leaving group to convert the 2′-3′ cyclic phosphate back to 2′OH and a 3′-phosphoryl group. Despite the similar mechanisms, the structural environments of His235 in nsp15 and the RNase A equivalent (His119) differ significantly, with the residues being ~8 Å apart and making several different hydrogen bonding interactions. These differences may provide an answer as to why nsp15 is much more sensitive to pH change compared to Rnase A [22]. What remains unclear is the contribution of Mn2+ to the reaction mechanism, particularly as an Mn2+ binding site has not been located in SARS-CoV-2 nsp15 [22].
Therapeutic interest of the protein
As previously mentioned, knockout studies on nsp15 have shown it is not essential for viral replication. Despite this, a nsp15 inhibitor could provide an effective treatment against SARS-CoV-2 by hampering its evasion and modulation of the innate immune response to help promote longer-lasting immunity. Targeting nsp15 is particularly interesting as nsp15 has no close human homologues [25], thereby potentially reducing harmful side effects. A number of biochemical assays have been performed on nsp15 to screen previously approved drugs and various libraries for inhibition of nsp15, as well as a number of in-silico studies to dock approved therapeutics to guide drug design efforts. A fragment screening study has also been performed that yielded 6 small molecule fragments.
Benzopurpurin B, C-473872 (CAS registry number: 331675-78-6), and Congo Red, as well as small molecular Rnase A inhibitors, have been shown to inhibit nsp15 activity and reduce infectivity of SARS-CoV in Vero cells [26] but further testing on SARS-CoV-2 nsp15 is required. Additionally, nsp15 has been screened against the ReFrame [27], Pandemic Response Box (Medicines for Malaria Venture (MMV) & Drugs for neglected disease initiative (DNDi)), and Covid Box drug repurposing libraries for 50% inhibition below concentration of 10 µM, identifying 23, 1, and 0 hits respectively from the libraries [25]. Two fluorescence resonance energy transfer (FRET) assays to determine the half-maximal inhibitory concentration (IC50) reduced the hits to 12 (11 in ReFrame, 1 in Pandemic Response Box), which were whittled down to 3 (Exebryl-1, Piroxantrone, and MMV1580853) after 9 were identified as false positives due to the production of reactive oxygen species such as H2O2, which destabilized protein in the assay. Ligand binding was assessed using high resolution mass spectrometry. Piroxantrone and MMV1580853 showed significantly weaker binding and ultimately no antiviral activity in SARS-CoV-2 assays. Exebryl-1 bound with an affinity constant Kd of ~12 µM per monomer in the first instance, with approximately four molecules binding to one monomer on average per 100 µM Exebryl-1; and molecular docking of Exebryl-1 against PDB entry 6XDH using an automated Qvina docking workflow [28] showed binding in a pocket close to and within the active site. Exebryl-1 demonstrated antiviral activity in three separate assays at concentrations over 10 µM. However, based on blood plasma levels in Sprague-Dawley rats after an oral dose of 100 mg/kg reaching only 9 µM after 1 hour, and dropping to 4 µM after 4 hours, Exebryl-1 is not expected to reach therapeutic levels in its current state [25].
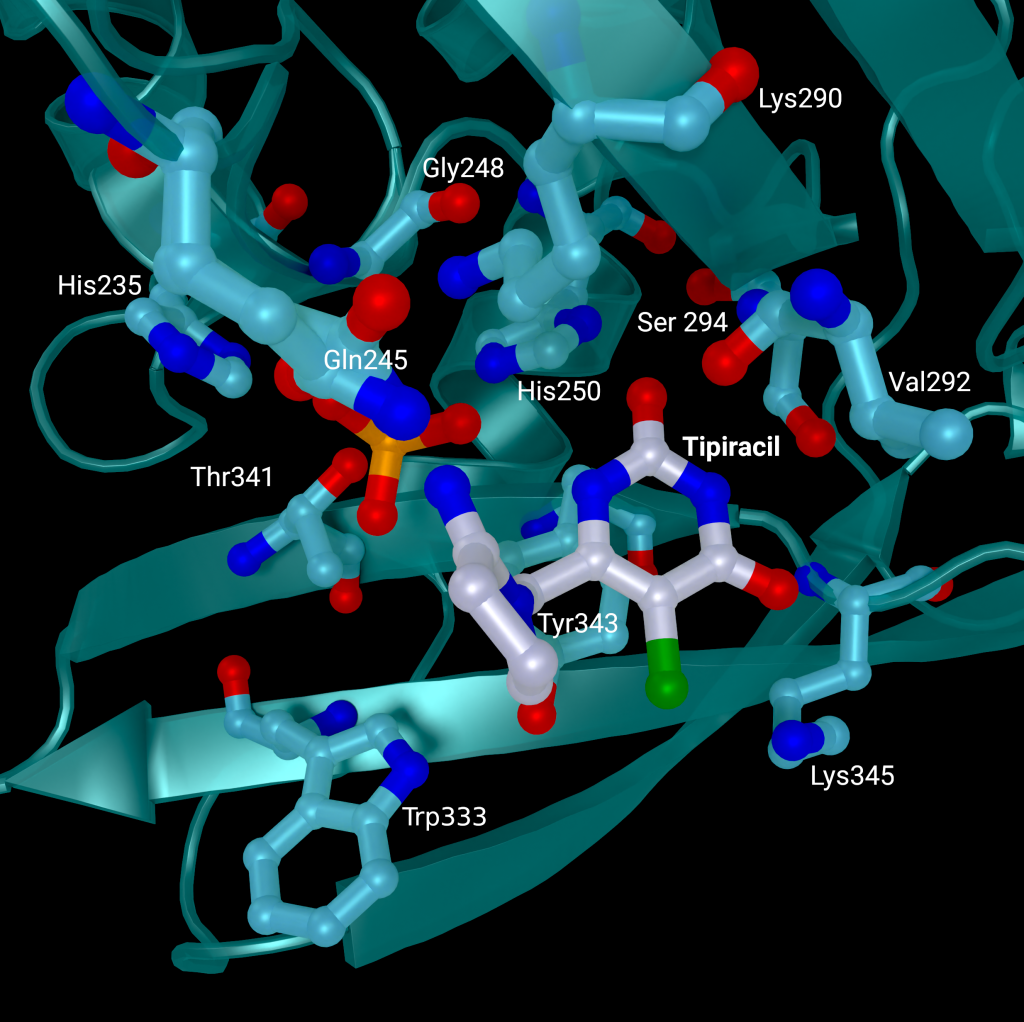
A repurposed colorectal cancer drug, Tipiracil, has been found to partially inhibit nsp15 activity in biochemical assays. However, the efficacy is greatly decreased in the presence of increased Mn2+ concentrations. A structure of nsp15 with Tipiracil interacting with the uridine binding pocket has also been solved (PDB entry: 6WXC), with its uracil ring stacking against Tyr341 and forming several hydrogen bonds with Ser294, Lys345, and His250 (Figure 5) as well as several interactions with other active site residues through water and phosphate molecules. The only unique interaction for this ligand is between the Iminopyrrolidin nitrogen of Tipiracil and Gln245 (Figure 5). Although not an immediate treatment option, the uracil derivative drug provides a potential scaffold for further SARS-CoV-2 nsp15 inhibitor development [23]. Based on Tipiracil binding at the active site a library of 85 flavinoid compounds were docked using the molecular mechanics/generalized Born surface area (MMGBSA) method and molecular dynamics with nsp15 (PDB entry 6WXC) as part of an in-silico study; but binding was found to be significantly weaker than Tipiracil in all cases [29].
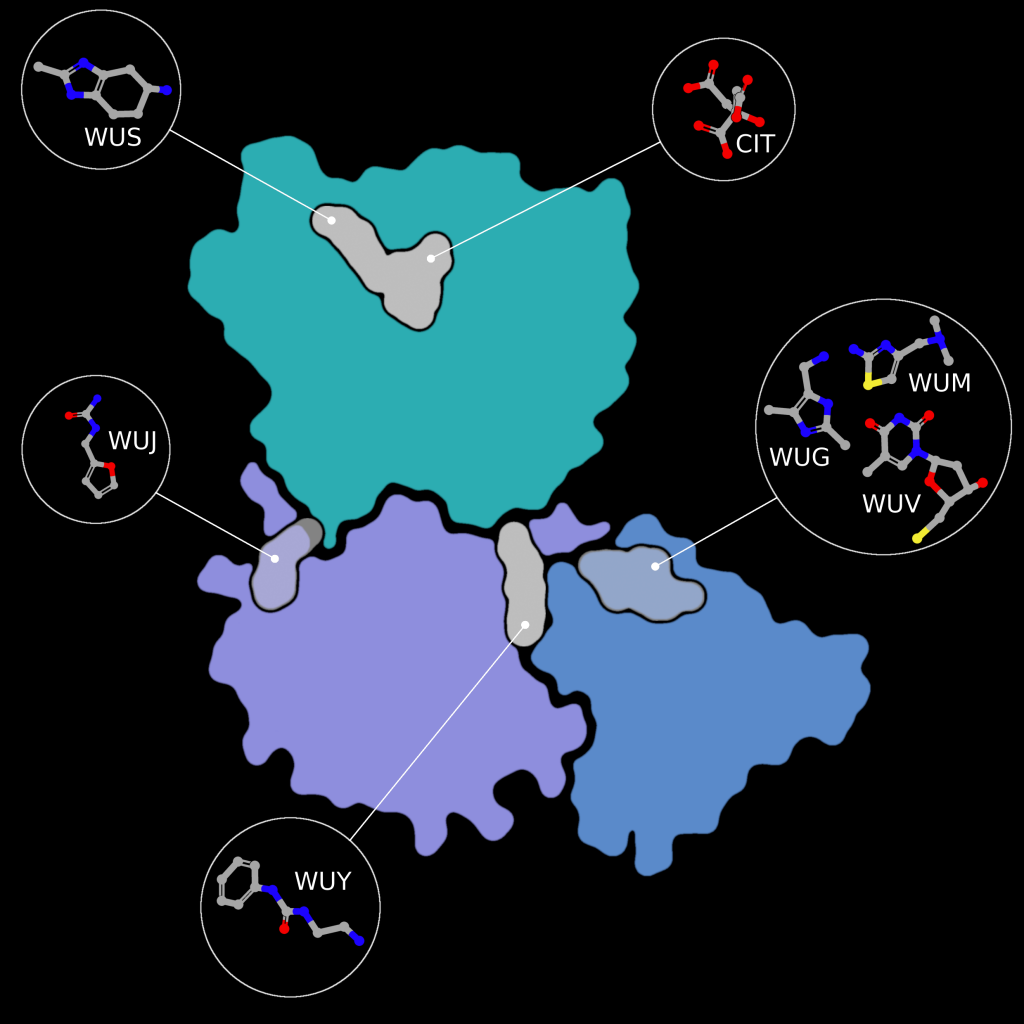
Fragment screens have been performed on nsp15, with six structures currently available in the PDB without an accompanying publication. In addition to the soaked fragments present in these structures, all show a citrate molecule bound to the catalytic NendouU domain (Figure 6, CIT), with one fragment bound adjacent to citrate (PDB entry 5S70, Figure 6, EN300-181428 (WUS)) through a stacking interaction with Trp333 and a hydrogen bond between the NO3 hydrogen of EN300-181428 and O5 of the citrate molecule. Four fragments are bound at the interface between the middle domain (Figure 6, purple) and the N-terminal oligomerisation domain (Figure 6, blue), including FUZS-5 (PDB entry 5S71, Figure 6, WUV) Z2889976755 (PDB entry 5S6X, Figure 6, WUG), BBL029427 (PDB entry 5S72, Figure 6, WUY), and PB2255187532 (PDB entry 5S6Z, Figure 6, WUM). Finally, BBL029427 (PDB entry 5S6Y, Figure 6, WUJ) is bound to a loop connecting beta strands in the middle domain. Unfortunately, the crystal packing in these structures prevents the formation of the active double-ringed hexamer structure using symmetry related molecules, making it difficult to assess how the fragments interact with the active hexamer. However, this monomeric crystal form could provide a starting point for the design of a drug to break up formation of the active hexamer by interfering with surfaces on the N-terminal oligomerization domain.
Molecular docking, all-atom molecular dynamics, and an assessment of absorption, distribution, metabolism, and excretion (ADME) properties have been carried out on PDB entry 6W01 using 15 scalarane sesterterpenes, compounds purified from Red Sea marine sponges with a variety of relevant pharmacological activities. to assess their efficacy as drug targets to inhibit nsp15 [30]. Eight compounds were found to have equivalent or better binding energies compared to the reference ligand, Benzopurpurin 4B. All eight compounds bound the C-terminal catalytic domain in the large shallow active site, forming polar interactions with the catalytic triad (His235, His250, and Lys290), interacting with Trp333 through π-stacking, and forming at least one hydrogen bond with Lys290 and further anchoring hydrogen bonds with Gly248 and/or Gln245 [30]. Two of the eight were used in all atom molecular dynamics simulations and showed good stability, high negative binding free energies, and scored well on ADME drug property predictions.
In-silico docking investigations of 32 phytochemicals from Asparagus racemous have also been performed on nsp15 (PDBID: 6W01). The top 5 ligands (Asparoside-C, Asparoside-F, Rutin, Asparoside-D, and Racemoside-A) bound at the C-terminal active site with binding free energy scores between ‒7.165 kcal/mol and ‒5.993 kcal/mol. Complexes of nsp15 and Asparoside-C, -F, and -D were subjected to further analysis by 100 ns molecular dynamics simulations, which found Asparoside-D and -F to have favorable binding interactions and better affinity than the control ligand Remdesivir [31]. 23 previously approved drugs have also been docked to nsp15, with three demonstrating high predicted binding affinities between ‒9.1 and ‒9.6 kcal/mol (Saquinavir, Aprepitant, and Valrubicin) [32]. However, the pocket Saquinavir, Aprepitant, and Valrubicin are docked to sites on the opposite side of the active site pocket which houses the catalytic triad, approximately 17 Å away. Barring an undetermined allosteric effect caused by this binding, which the paper makes no mention of, further development of these drug targets “…modifying them to fit to the SARS-CoV-2 nsp15 active site pocket precisely” needs to be rethought as the active site has not been targeted in the first instance.
Complementary knowledge
The enzymatic activity of nsp15 and its crystal structure have been demonstrated, but the exact role in viral replication remains unclear. SARS-CoV nsp15 has been shown to co-localize with replicating RNA [33] around the nucleus as well as nsp8 and nsp12 from the replication/transcription complex in in situ studies [34], in the presence and absence of RNA. It was also shown that SARS-CoV nsp15 does not co-localise with the M protein [34]. Yeast two-hybrid screens and glutathione S-transferase (GST) pulldown assays have also identified nsp8 and nsp12 as potential binding partners to SARS-CoV nsp15 [35].
Furthermore, nsp15 has demonstrated a strong inhibitory effect on interferon (IFN) production and interferon regulatory factor 3 nuclear localization in in-vitro co-expression assays against the Cantell strain of Sendai virus with nsp13, nsp14, and accessory protein ORF6 [36]. However, interferon antagonization in in-vitro conditions is not necessarily representative of real infection, individual protein expression levels can vary greatly compared to overexpression studies and altered localization can have a significant effect[36]. The individual contribution or mechanism of nsp15 interferon inhibition is not discussed by Yuen et al 2020 in this study. Overall SARS-CoV-2 appears less effective at suppressing interferon signaling compared to SARS-CoV due to the loss of SARS-CoV-2 papain-like protease (PLpro) as an interferon antagonist [36]. Reverse genetic studies (analysis of a resulting phenotype following genetic engineering) have suggested that ORF6 is the major player in interferon suppression instead [37]. However, SARS-CoV-2 ORF6 is also less conserved between SARS-CoV and SARS-CoV-2 at only 69% sequence identity and only 4 of 10 key amino acids identified from SARS-CoV ORF6 being present in SARS-CoV-2 ORF6 [36].
It has been shown that nsp15 activity is highly dependent on the presence of Mn2+ ions, showing greatly reduced activity in the presence of Mg2+ ions. In the presence of Mn2+ nsp15 was able to cleave all four uridine sites in an eicosamer, a 20-subunit oligomer consisting of 5′GAACU↓CAU↓GGACCU↓U↓GGCAG3′, with no preference for sequence and increased cleavage rate with rising metal ion concentration [23]. This is particularly interesting as Mn2+ enhances activity in SARS-CoV nsp15, but protein activity does not depend on the presence of Mn2+, and no metal binding sites have been identified in coronavirus structures to date [18]. Considering SARS-CoV-2 nsp15 shares 88% sequence identity with SARS-CoV nsp15, and all active site residues are conserved, SARS-CoV 2 nsp15’s dependence on Mn2+ is a significant difference between the enzymes. Further to this, nsp15 alone is promiscuous, cutting any uridine sites in RNA, but becomes site-specific when in complex with nsp8 and nsp12 and leaves uridine tails between 5 and 10 bases long [16].
A library of 5000 small molecule compounds has been screened against nsp15 for inhibition of nuclease activity, with twelve compounds showing potential as antiviral treatments in a fluorescent biochemical kinetic screen. Further analysis using a gel-based assay found only one compound, NSC95397, able to inhibit nuclease activity at a concentration of 10 µM. However, tests on SARS-CoV 2 infected VERO E6 cells found the compound toxic at concentrations above 10 µM and ineffective at inhibiting viral growth at lower concentrations [38].
A fluorescence resonance energy transfer (FRET) assay has been performed to measure nsp15 activity on a 6-mer oligonucleotide (5′-AAAUAA) with a 5′-fluorescein and 3′-TAMRA label [21,22]. Activity is measured through an increase in fluorescence caused by the removal of the 5′-TAMRA label. Nsp15 activity was confirmed for the wild-type protein and abolished in H235A and H250A mutants [22]. FRET analysis was paired with liquid chromatography electrospray ionization mass spectrometry to demonstrate that nsp15 3′RNA products show a preference for accumulation of 2′-3′ cyclic phosphate (80%) compared to 3′-phosphate, a significant difference compared to RNAse A which generates a 2’-3’ cyclic phosphate which is then hydrolysed to a 3’-phosphate.
Summary
SARS-CoV 2 nsp15 is an RNA uridylate‐specific Mn2+-dependent [3] endoribonuclease from the nidoviral endoU (NendoU) family, which acts on single-stranded and double-stranded RNA to help SARS-CoV-2 evade detection by the innate immune response. Knockout studies have demonstrated that nsp15 is not essential for viral replication, but numerous studies have shown a reduction in viral titre and virulence in nsp15-deficient SARS-CoV-2 when studied in the presence of an effective immune response.
The sequence of nsp15 is highly conserved between SARS-CoV-2, SARS-CoV, MERS-CoV, and HCoV-229E, as is the fold of the monomer and active hexamer. The monomer consists of three domains, the N-terminal oligomerisation domain, a middle domain, and the NendoU catalytic domain which houses the active site. The active site is a shallow groove made up of six key residues (His235, His250, Lys290, Thr341, Tyr343, and Ser294). A series of structures with different catalytic intermediates have been solved and the reaction mechanism is predicted to act in a similar manner to the well-studied RNaseA enzyme. However, nsp15’s dependence on manganese, where RNase A’s activity is metal independent, throws some aspersions on this theory.
Three in-silico drug screeningstudies have been performed on nsp15, two using 6W01 and one using 6WXC as the protein models. 6W01 is a citrate bound nsp15 structure solved to 1.9 Å resolution, with acceptable data processing and refinement statistics overall, the only minor concern is that 5% of the residues in both chains show one issue with their geometry, and a small subset of that 5% show an issue in their fit to the electron density. 6WXC is a Tipiracil bound nsp15 structure solved to 1.85 Å resolution, it faces a similar minor problem to 6W01 with 7% of residues in both chains showing one issue with their geometry but with fewer electron density fit outliers. Use of either model should present no major stumbling blocks for simulation studies.
Discussion & Outlook
Nsp15 has been one of the lesser explored proteins compared to other SARS-CoV 2 proteins, such as the main protease and the papain-like protease, which have undergone extensive in-silico drug design studies through a number of large collaborative efforts between universities, synchrotrons, and other organizations [39–45] to feed into the COVID Moonshot project [46]. Overall, the structural work on nsp15 has been sound and all available models could provide a good starting structure for computational drug design. A series of structures with catalytic intermediates suggests a mechanism akin to RNase A, however, the dependence of nsp15 on Mn2+ suggests a departure from this mechanism as RNase A’s mechanism is metal independent. Follow up in-silico studies (described above) were based on well validated models with acceptable statistics for the resolution the structures were solved at, although none have yet pointed to a viable lead compound for clinical application. Nsp15 not being essential for viral replication makes it a much less desirable target for structure-based drug design compared to other essential viral proteins. However, the impact of nsp15 on SARS-CoV-2’s virulence by repressing the innate immune response shows a potential avenue to weaken SARS-CoV-2 through inhibition of nsp15 to allow the immune system to fight off infection before it becomes more severe.
Acknowledgements
This work was supported by the German Federal Ministry of Education and Research [grant no. 05K19WWA], Deutsche Forschungsgemeinschaft [project TH2135/2-1]. The authors would also like to thank Johannes Kaub and Rosemary Wilson for support and discussion. All figures are courtesy of the Coronavirus Structural Task Force (insidecorona.net), who retains copyright for the text and the figures..
[1] Kim Y, Jedrzejczak R, Maltseva NI, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Science. 2020;29:1596–1605.
[2] Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192.
[3] Ivanov KA, Hertzig T, Rozanov M, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci U S A. 2004;101:12694–12699.
[4] Naqvi AAT, Fatima K, Mohammad T, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165878.
[5] Bhardwaj K, Sun J, Holzenburg A, et al. RNA Recognition and Cleavage by the SARS Coronavirus Endoribonuclease. J Mol Biol. 2006;361:243–256.
[6] Deng X, Baker SC. An “Old” protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses. Virology. 2018;517:157–163.
[7] Snijder EJ, Decroly E, Ziebuhr J. Chapter Three - The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. In: Ziebuhr J, editor. Advances in Virus Research [Internet]. Academic Press; 2016 [cited 2022 Jan 7]. p. 59–126. Available from: https://www.sciencedirect.com/science/article/pii/S0065352716300471.
[8] Nga PT, Parquet M del C, Lauber C, et al. Discovery of the First Insect Nidovirus, a Missing Evolutionary Link in the Emergence of the Largest RNA Virus Genomes. PLOS Pathogens. 2011;7:e1002215.
[9] Lauber C, Ziebuhr J, Junglen S, et al. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch Virol. 2012;157:1623–1628.
[10] Tomasello G, Armenia I, Molla G. The Protein Imager: a full-featured online molecular viewer interface with server-side HQ-rendering capabilities. Bioinformatics. 2020;36:2909–2911.
[11] Deng X, Hackbart M, Mettelman RC, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. PNAS. 2017;114:E4251–E4260.
[12] Kindler E, Gil-Cruz C, Spanier J, et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLOS Pathogens. 2017;13:e1006195.
[13] Volk A, Hackbart M, Deng X, et al. Coronavirus Endoribonuclease and Deubiquitinating Interferon Antagonists Differentially Modulate the Host Response during Replication in Macrophages. Journal of Virology [Internet]. 2020 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/JVI.00178-20.
[14] Kato H, Takeuchi O, Sato S, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105.
[15] Mandilara G, Koutsi MA, Agelopoulos M, et al. The Role of Coronavirus RNA-Processing Enzymes in Innate Immune Evasion. Life (Basel). 2021;11:571.
[16] Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc Natl Acad Sci U S A. 2020;117:8094–8103.
[17] Deng X, Geelen A van, Buckley AC, et al. Coronavirus Endoribonuclease Activity in Porcine Epidemic Diarrhea Virus Suppresses Type I and Type III Interferon Responses. Journal of Virology [Internet]. 2019 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/JVI.02000-18.
[18] Ricagno S, Egloff M-P, Ulferts R, et al. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. PNAS. 2006;103:11892–11897.
[19] Joseph JS, Saikatendu KS, Subramanian V, et al. Crystal Structure of a Monomeric Form of Severe Acute Respiratory Syndrome Coronavirus Endonuclease nsp15 Suggests a Role for Hexamerization as an Allosteric Switch. Journal of Virology [Internet]. 2007 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/JVI.02817-06.
[20] Bhardwaj K, Palaninathan S, Alcantara JMO, et al. Structural and Functional Analyses of the Severe Acute Respiratory Syndrome Coronavirus Endoribonuclease Nsp15*. Journal of Biological Chemistry. 2008;283:3655–3664.
[21] Zhang L, Li L, Yan L, et al. Structural and Biochemical Characterization of Endoribonuclease Nsp15 Encoded by Middle East Respiratory Syndrome Coronavirus. Journal of Virology [Internet]. 2018 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/JVI.00893-18.
[22] Pillon MC, Frazier MN, Dillard LB, et al. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat Commun. 2021;12:636.
[23] Kim Y, Wower J, Maltseva N, et al. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun Biol. 2021;4:1–11.
[24] Saramago M, Costa VG, Souza CS, et al. The nsp15 Nuclease as a Good Target to Combat SARS-CoV-2: Mechanism of Action and Its Inactivation with FDA-Approved Drugs. Microorganisms. 2022;10:342.
[25] Choi R, Zhou M, Shek R, et al. High-throughput screening of the ReFRAME, Pandemic Box, and COVID Box drug repurposing libraries against SARS-CoV-2 nsp15 endoribonuclease to identify small-molecule inhibitors of viral activity. PLOS ONE. 2021;16:e0250019.
[26] Ortiz-Alcantara J, Bhardwaj K, Palaninathan S, et al. Small molecule inhibitors of the SARS-CoV Nsp15 endoribonuclease. Virus Adaptation and Treatment. 2010;2:125–133.
[27] Janes J, Young ME, Chen E, et al. The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. PNAS. 2018;115:10750–10755.
[28] Alhossary A, Handoko SD, Mu Y, et al. Fast, accurate, and reliable molecular docking with QuickVina 2. Bioinformatics. 2015;31:2214–2216.
[29] Mishra GP, Bhadane RN, Panigrahi D, et al. The interaction of the bioflavonoids with five SARS-CoV-2 proteins targets: An in silico study. Comput Biol Med. 2021;134:104464.
[30] Elhady SS, Abdelhameed RFA, Malatani RT, et al. Molecular Docking and Dynamics Simulation Study of Hyrtios erectus Isolated Scalarane Sesterterpenes as Potential SARS-CoV-2 Dual Target Inhibitors. Biology (Basel). 2021;10:389.
[31] Chikhale RV, Sinha SK, Patil RB, et al. In-silico investigation of phytochemicals from Asparagus racemosus as plausible antiviral agent in COVID-19. J Biomol Struct Dyn. 2021;39:5033–5047.
[32] Mahmud S, Elfiky AA, Amin A, et al. Targeting SARS-CoV-2 nonstructural protein 15 endoribonuclease: an in silico perspective. Future Virol. :10.2217/fvl-2020–0233.
[33] Shi ST, Schiller JJ, Kanjanahaluethai A, et al. Colocalization and Membrane Association of Murine Hepatitis Virus Gene 1 Products and De Novo-Synthesized Viral RNA in Infected Cells. Journal of Virology [Internet]. 1999 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/JVI.73.7.5957-5969.1999.
[34] Athmer J, Fehr AR, Grunewald M, et al. In Situ Tagged nsp15 Reveals Interactions with Coronavirus Replication/Transcription Complex-Associated Proteins. mBio [Internet]. 2017 [cited 2022 Jan 6]; Available from: https://journals.asm.org/doi/abs/10.1128/mBio.02320-16.
[35] Imbert I, Snijder EJ, Dimitrova M, et al. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148.
[36] Yuen C-K, Lam J-Y, Wong W-M, et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 9:1418–1428.
[37] Schroeder S, Pott F, Niemeyer D, et al. Interferon antagonism by SARS-CoV-2: a functional study using reverse genetics. The Lancet Microbe. 2021;2:e210–e218.
[38] Canal B, Fujisawa R, McClure AW, et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp15 endoribonuclease. Biochem J. 2021;478:2465–2479.
[39] Cantrelle F-X, Boll E, Brier L, et al. NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment. Angewandte Chemie International Edition. 2021;60:25428–25435.
[40] Newman JA, Douangamath A, Yadzani S, et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat Commun. 2021;12:4848.
[41] Zhao Y, Du X, Duan Y, et al. High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors. Protein Cell. 2021;12:877–888.
[42] Ma C, Sacco MD, Xia Z, et al. Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay. ACS Cent Sci. 2021;7:1245–1260.
[43] Douangamath A, Fearon D, Gehrtz P, et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat Commun. 2020;11:5047.
[44] Ahmad S, Abdullah I, Lee YK, et al. Extensive Crystallographic Fragment-Based Approach to Design SARS CoV2 3CLpro Main Protease Inhibitors and Related Metadata. 2021 [cited 2022 Jan 6]; Available from: https://chemrxiv.org/engage/chemrxiv/article-details/60c753c8469df4c73ef44e13.
[45] Günther S, Reinke PYA, Fernández-García Y, et al. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372:642–646.
[46] Consortium TCM, Achdout H, Aimon A, et al. Open Science Discovery of Oral Non-Covalent SARS-CoV-2 Main Protease Inhibitor Therapeutics [Internet]. 2021 [cited 2022 Jan 6]. p. 2020.10.29.339317. Available from: https://www.biorxiv.org/content/10.1101/2020.10.29.339317v2.
