Für diesen Artikel steht leider keine deutsche Übersetzung zur Verfügung.
An important drug target
In the first part of this series we compared the protein nsp3 from SARS-CoV and SARS-CoV-2 by sequence. Now we delve deeper into the differences between these two proteins and follow through by analyzing the structure of one domain of nsp3 in particular: papain-like protease. This domain is a very relevant drug target because of its ability not only to cleave the polyprotein, but also remove some of the post-translational modification our cells use to fight these viruses. Without papain-like protease, the virus would be unable to spread COVID-19.
Like the entire nsp3 protein, the papain-like-protease (Pl2pro) domain is localized close to the endoplasmic reticulum’s (ER) membranes. The transmembrane domains hold it in place while the majority of the protein protrudes out of the ER membrane into the cytoplasm.[1]
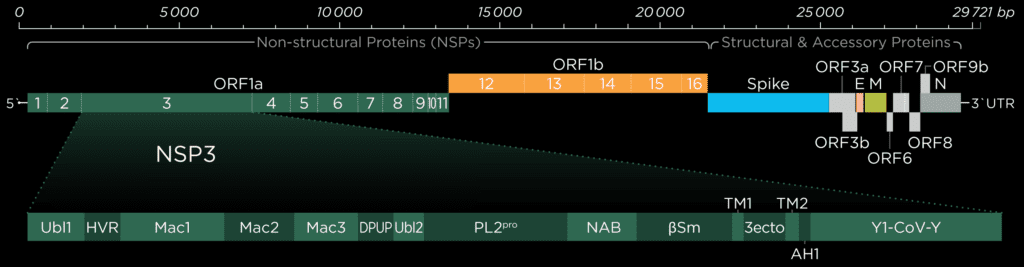
Ubiquitin-like-domain 2
We cannot discuss the Pl2pro domain without its little neighbor, which has been speculated to influence protease domain functionality.
In ubiquitin-specific proteases, the function of comparable Ubl2 domains is attributed to substrate recruitment or an increase in catalytic efficency. Ubiquitin-like-domain 2 (Ubl2) is the domain residing directly adjacent to the N-terminus of the Pl2pro catalytic domain. These ubiquitin-like domain seems to be more conserved compared to Ubl1 in different coronavirus species.[2]
If, in SARS-CoV and Murine coronavirus (MHV), Ubl2 is removed, Pl2pro loses its structural integrity. In addition, Pl2pro is then no longer able to act as an Interferon (IFN) antagonist (see below). However, some studies suggest that the Ubl2 domain in MERS-CoV might not be as essential as originally thought and in cell-based studies of this virus, Pl2pro could retain some of its enzymatic functions without the Ubl2 domain.[3]
To date, several inconsistent roles of Ubl2 were reported, and its exact function and inner workings remain enigmatic. This is being highlighted the fact that there are significant differences between the coronaviruses, and as a consequence, we need to exercise caution in applying our findings to SARS-CoV-2.
Combating the Host's Immune System
In the family of coronaviridae, viruses with either one or two Plpro domains can be found, with SARS-CoV and SARS-CoV-2 only having one. Confusingly, this single domain is however still called Pl2pro, even if it is the only papain-like protease domain in the viral genome.
Pl2pro cleaves the polyprotein from nsp1 (leader protein) up to nsp3. While Pl2pro cuts between nsp1-( ELNGG↓AV)-nsp2-( RLKGG↓AP)-nsp3-( SLKGG↓KI)-nsp4, the nsp5 (3c-like protease) cleaves the rest of the polyprotein. [2] The cysteine protease Plpro is similar to human ubiquitin-specific-protease (USP) in that it adopts a right-hand fold with "thumb", "palm" and "finger" subdomains.
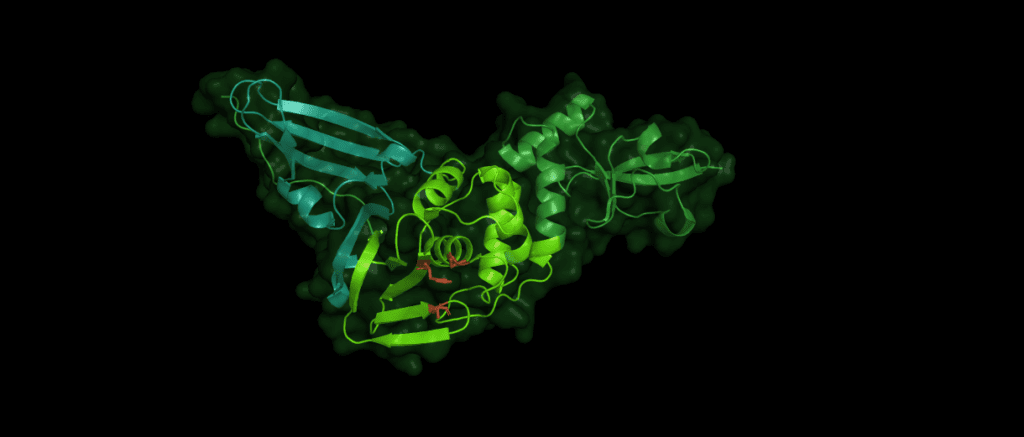
Despite the variations of Pl2pro in different coronaviridae, the same catalytic motif of three amino acid residues is essential for the stability and proteolytic activity of the domain: Cys112 is located in the thumb, His273 and Asp287 are located in the palm subdomain. (The numbers identifying these residues can vary between species.)
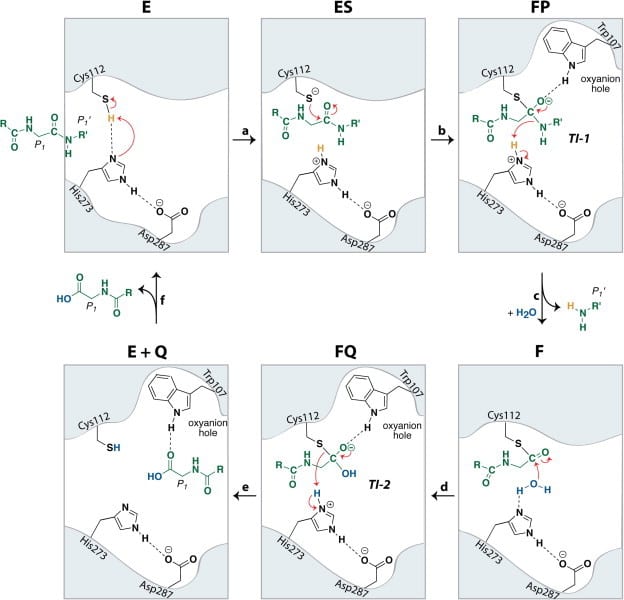
In addition, Pl2pro has deubiquitinating and deISGylating (removal of ISG15 from target proteins) abilities.[4] Both ubiquitin and ISG15 regulate facets of the immune response and through their removal Pl2pro poses as an antagonist to the human immune response. They can stimulate the production of cytokines, chemokines and other IFN-stimulated gene products which have antiviral properties. [6] ISG 15 is an ubiquitin-like modifier composed of two ubiquitin-like folds that has an essential role in marking newly synthesized proteins during the antiviral response.[3] Post-translational modification by ubiquitin and interferon-stimulating gene 15 (ISG15) is reversed by isopeptide bond hydrolysis. Figure 3 shows a proposed mechanism for the cleaving of isopeptide bonds by SARS-CoV.
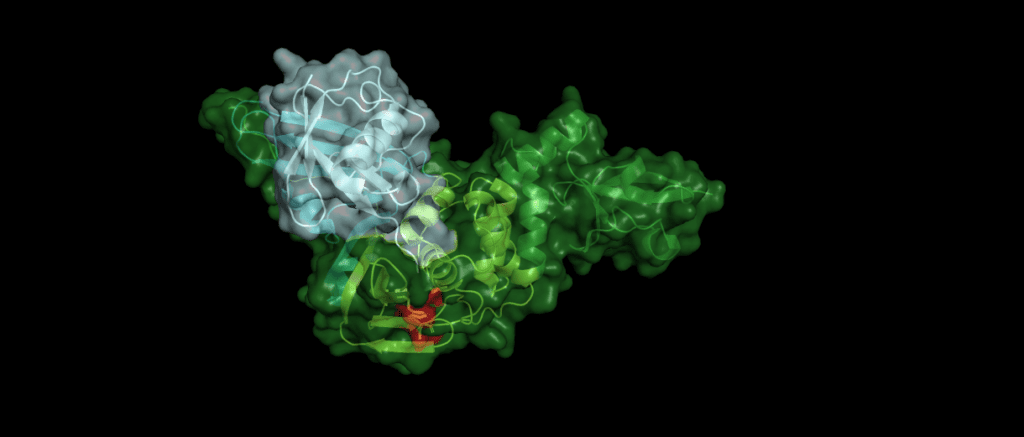
An example
Toll-like receptors (TLRs) are an important part of the machinery of the human immune response, which recognizes the pathogen-associated molecular patterns. The ability of the host cell to transduce the so-called Toll-like receptor 7 (TLR7) mediated immune response is diminished (Fig. 5) by Pl2pro as it removes Lys63-linked-ubiquitin from the TNF receptor associated factors TRAF3 and TRAF6. [5]
In addition, SARS-CoV can hamper the antiviral activities of interferon. The Pl2pro domain inhibits in combination with a transmembrane (TM) domain the STING mediated activation of interferon expression. PL2pro-TM interacts with TRAF3, TBK1, IKKε, STING and IRF3, the key components assembling a regulatory complex for activation of IFN expression.[5]
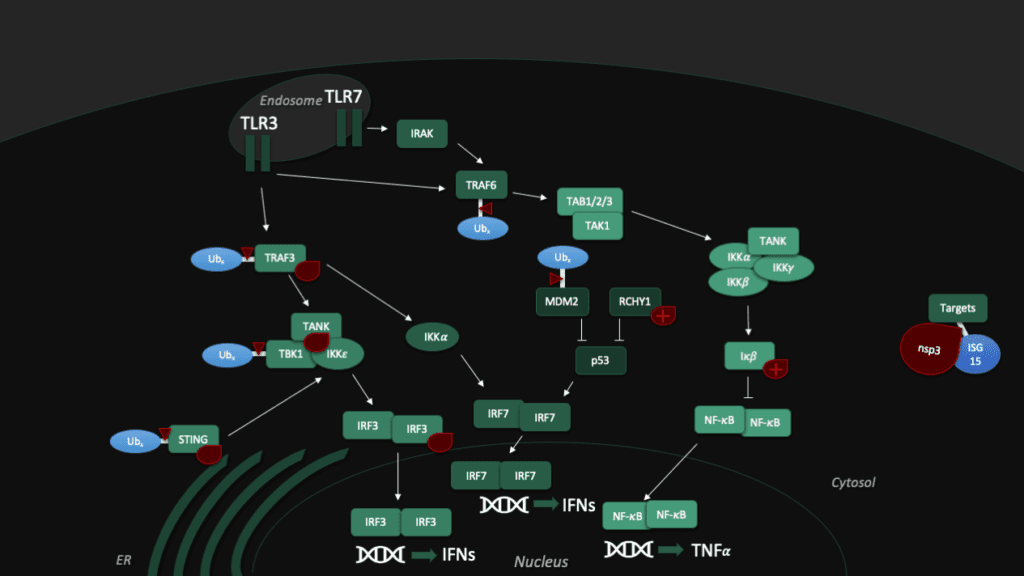
Another tool to fight the coronavirus in human cells is the "guardian of the genome", p53. The tumor supressor protein p53 impedes the replication of SARS-CoV, though the virus fights back with Pl2pro, which binds a p53 degradation stimulator named "RING finger and CHY zinc finger domain-containing protein 1" (or short: RCHY1). Enhanced by the Macro somains in NSP3, this binding enhances the stability of RCHY1 and hence promotes the degradation of p53. In addition, Pl2pro blocks another crucial cellular defense mechanism: The NF-κB pathway, which regulates immune responses to infections. SARS-CoV Pl2pro can stabilize IκBα, an inhibitor of NF-κB.[3]
Although all Pl2pro in different coronaviridae suppress the immune response, the targets differ between various species. For example, SARS-CoV Pl2pro preferentially processes Lys48 linked poly-ubiquitin chains, which are markers for proteasome degradation. MERS, on the other hand, shows no differences in effectivity between Lys48 and Lys63 linked di-Ubq chains. Lys63-linked chains are related to signal transduction cascades of the host immune system. Studies have shown that specificity among Pl2pro for Ubiquitin and ISG15 substrates can be altered with as little as a single amino acid change.[6] However, even though there are differences, for SARS-CoV-2, it is likely that at least some of the functions are similar.
Structural comparison
In order to predict Pl2Pro function for the novel Coronavirus SARS-CoV-2, we start by aligning their sequence like we did in the first part of this series to comapare the sequence with the one from SARS-CoV-2. Both domains share a similarity of 82.8% over the length of 313 amino acids. However, this time, we go for a more detailed analysis of the 54 individual differences, which are:
T3R N14I V20V N48N H49S V56Y D60N E66V D75T S77P P95Y G99N S114A V115T L116A L119T E123I K125L P129P A134D A143E N155C H170S L171Y Q173F S179D K181C C191T T195Q T196Q G200K N214E L215Q K216F G218K I221Q C225T D228K A229Q Y232K F240P Y250Q L252E Q254K G255H C259T E262S H274K K278S I284C L289L S293S T300I S308N
(The first letter refers to SARS-CoV, and the second to the amino acid residue in SARS-CoV-2.)
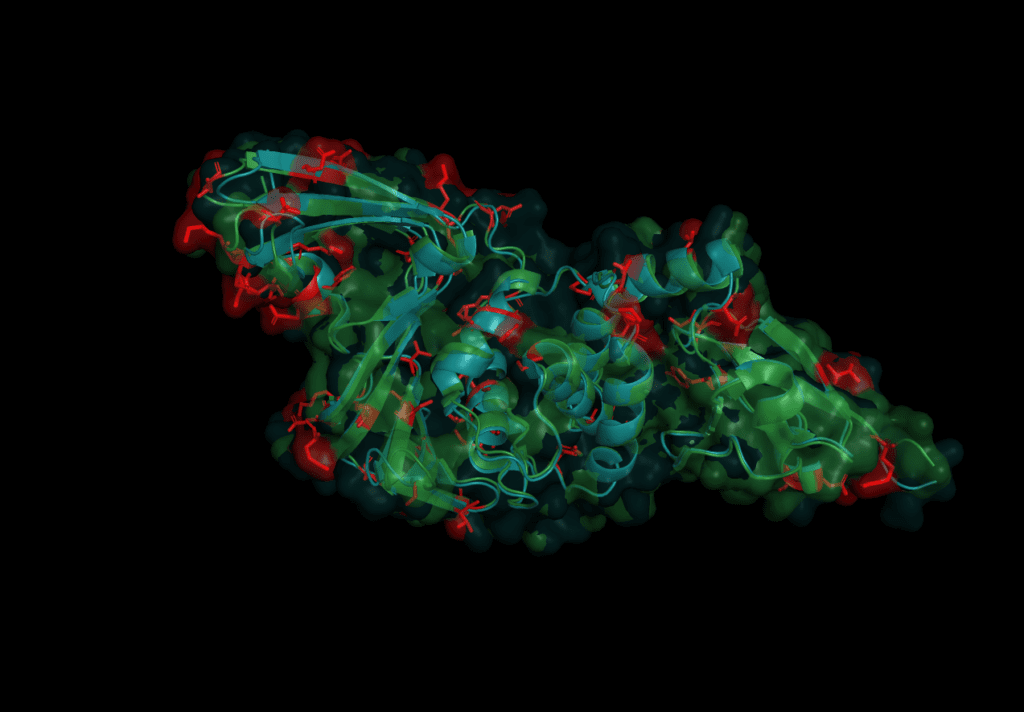
The mutations are evenly spread over the protein. None of the catalytic triad (Cys 112, His 273, Asp287) are changed as is to be expected given their conservation in all other coronaviruses. On further investigation, however, in the motif which interacts with ubiquitin six sites are different: S170T, Y171H, F216L, Q195K, T225V, and K232Q. Earlier studies concluded that the mutation of position 232 from Glutamine to Lysine increases the affinity for ubiquitin at the expense of the de-ubiquitination effectiveness.[6] The kinetics of SARS-CoV-2 nsp3 Pl2pro were studied to test if the protease domain of nsp3 has a reduced effectiveness in binding ubiquitin compared to nsp3 from SARS-CoV, MERS-CoV.
All three Pl2Pro variants cleave more ISG15 than ubiquitin. SARS-CoV has the fastest kinetics of the three viruses. And, the slower kinetics of SARS-CoV-2 resemble those of MERS-CoV rather more than SARS-CoV, having a 10 times higher turnover rate (kcat) as a deISGylase than as a deubiquitase.[6]
Besides the kinetics, the Pl2pro’s affinity for different poly-ubiquitin linkage sites was measured. The result shows that while SARS-CoV-2 can cut K48-Ub linked polyproteins, it seems to lack an ability to cut other polyubiquitin chains. Those K48-Ub linked polyproteins are cleaved at a slower rate than by SARS-CoV. In this regard, SARS-CoV-2 distinguishes itself from MERS-CoV which has the ability to cleave K63-linkages. It is suggested that the decrease in deubiquitinase effectiveness may not be irrelevant, but could lead to the often-mild symptoms that are a factor in why SARS-CoV-2 has been able to evade our efforts in quarantine. But this is mere speculation and a lot more research is needed to resolve the matter.[6]
PL2pro as a drug target
Pl2pro was a potential drug target early on in SARS-CoV-2 research. Hilgenfeld et al. name two major challenges we have to overcome to find a drug targeting Pl2pro. One is that the binding sites are tailor-made to bind glycine residues. Also, this very specific binding motif is rather ubiquitious in our cells. These two problems make it difficult to find an inhibitor which fits and is specific to Pl2pro. However, scientists found a weak spot: a loop called Blocking Loop 2 (BL2) regulates substrate binding and may be a promising target to inhibit PL2pro.[2] Naphthalene based inhibitors, which were earlier proposed to inhibit the BL2 of SARS-CoV, were shown to also inhibit SARS-CoV-2 Pl2pro, in particular an inhibitor called GRL-0617.[6]
For in-silico drug development, it might be prudent to choose high-resolution structures which already have a ligand or inhibitor bound, such as 6yva, 6wuu, 6wx4 or 6yaa. Technically speaking, 6wrh, albeit being a mutant, is one of the highest-quality structures available for SARS-CoV-2 Pl2pro.
In fact, a lot of research is still required to consolidate our understanding of this protein and its domains. In spite of that, we are making progress in our endeavor to fight this virus - and every step we take is one more to win this fight.
Sources
[1] Báez-Santos YM, St John SE, Mesecar AD. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21-38. doi:10.1016/j.antiviral.2014.12.015, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5896749/
[2] Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58-74. doi:10.1016/j.antiviral.2017.11.001, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7113668/
[3] Clasman JR, Báez-Santos YM, Mettelman RC, O'Brien A, Baker SC, Mesecar AD. X-ray Structure and Enzymatic Activity Profile of a Core Papain-like Protease of MERS Coronavirus with utility for structure-based drug design. Sci Rep. 2017;7:40292. Published 2017 Jan 12. doi:10.1038/srep40292, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5228125/
[4] Lei J, Hilgenfeld R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 2017;591(20):3190-3210. doi:10.1002/1873-3468.12827, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7163997/
[5] Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5(5):369-381. doi:10.1007/s13238-014-0026-3, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3996160/
[6] Freitas BT, Durie IA, Murray J, et al. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease [published online ahead of print, 2020 Jun 4]. ACS Infect Dis. 2020;acsinfecdis.0c00168. doi:10.1021/acsinfecdis.0c00168, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7274171/
