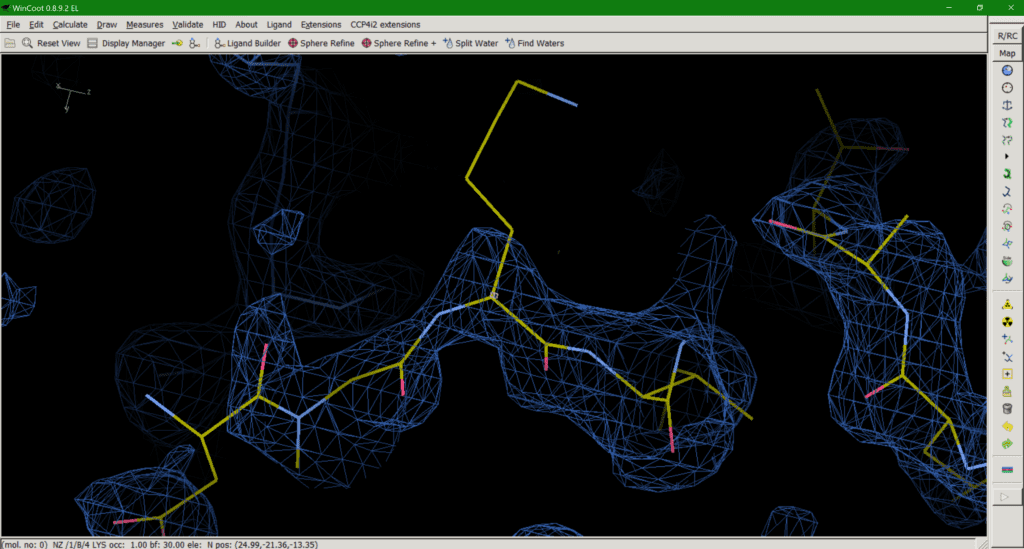
Crystallography has a problem. Some amino acid side chains in our structures simply can’t be seen in our maps (Fig. 1). Crystallographic maps represent many protein molecules in a crystal lattice, thousands of copies of the same molecule averaged over measurement time and unit cells. So, what happens with inherently flexible regions of our protein? The average of many different conformations leaves us with no map to guide us in modelling our side chain. So, what is the best way to deal with this as a model builder?
A passionate discussion within the Task Force has resulted in the following options for dealing with this situation:
- Set the occupancy of the unresolved atoms to 0
- Leave the atoms at full occupancy and allow the B-factors to inflate
- Trim the side chains to what can be resolved by the density
- Mutate the residue to a Proline, set your computer on fire, and walk away laughing maniacally.
Just to be clear, option four should only be considered in the direst of circumstances. Please consider options one to three before resorting to proline and fire, and even then, only with a computer you own. With that said, what is the best option? Sadly, none are ideal solutions to the problem so let’s discuss.
Option 1 can be misleading as the residue appears to be present in the model (Fig. 2), despite there being no experimental evidence for it, until you check the occupancy or load the corresponding map with your model which will tell you otherwise. An occupancy of zero also adds no useful information to the model and may even exclude atoms in this position, like opening the airlock and sending it flying out into the vacuum of space.
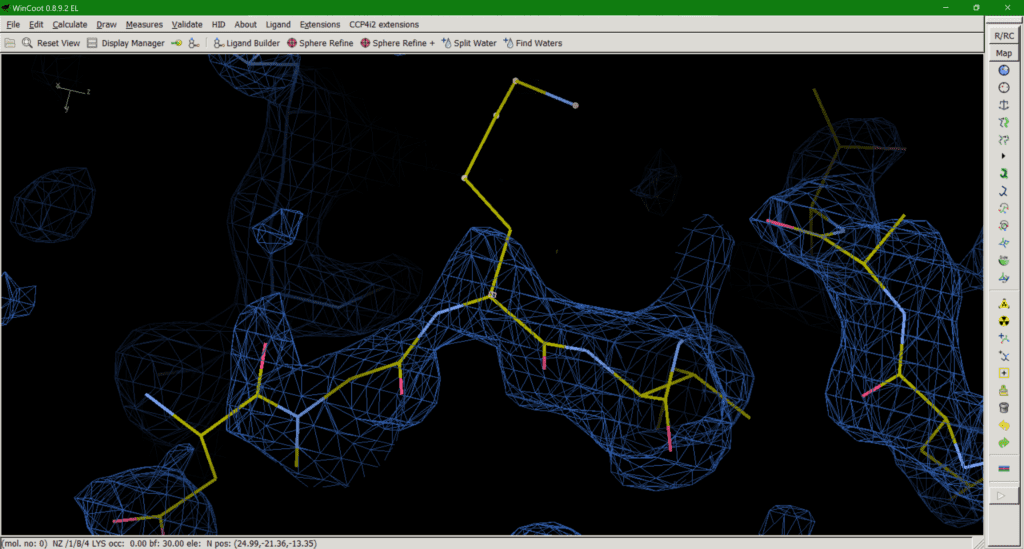
Option 2 is effectively the opposite of option 1, providing a full occupancy side chain in a sensible rotamer conformation and accept the resulting phase bias*. However, this can be equally misleading if the downstream user doesn’t check the B-factors of the sidechain, which will be very large, as they represent not only (smaller) displacement but (larger) disorder. In addition, allowing the B-factor to “explode” is not always an effective way to deal with this problem, as strong negative peaks can still be observed around the side chain in some cases. Another argument for maintaining an occupancy of 1 is that the protein sequence tells us a certain amino acid is present at a position, unless evidence of chemical clipping has been provided (mass spec, for example). Therefore, the atoms must be present in the protein so should be included in the model for the B-factors to deal with the physics of the situation. Options 1 and 2 both have the advantage of providing a complete set of atoms for downstream use in molecular modelling.
*During refinement our model will always bias the phase calculation which gives us our maps. Ideally, we would like out model to maximally affect the phases when we are confident our model is correct and minimally affect the phases when we are less confident. So, an occupancy of 1 (high confidence) where we observe no peaks in our map (low confidence) will lead to what we call phase bias. This can work both ways by underestimating the contribution of our model by setting the occupancy to 0 (option 1).
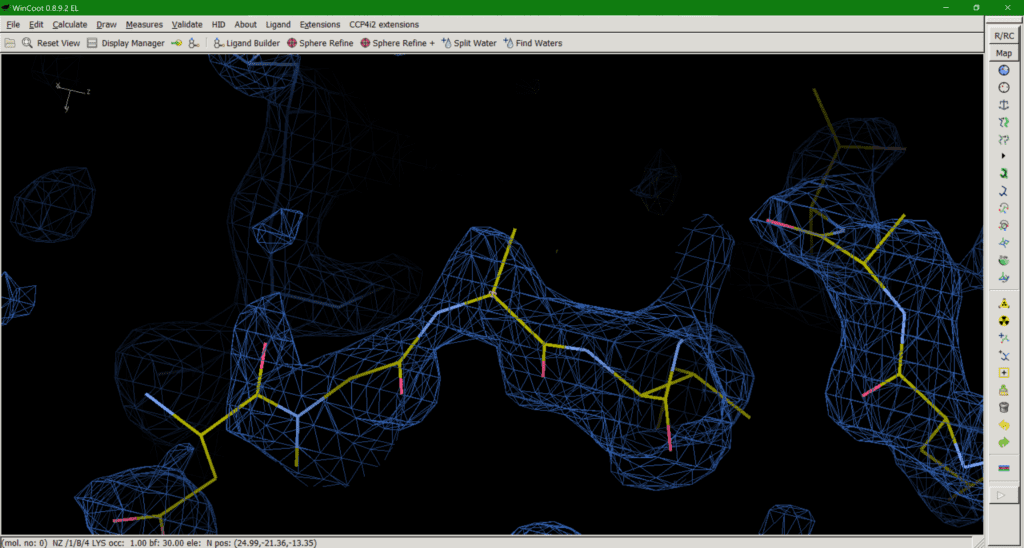
This brings us onto option 3: trimming down the side chain to what we can in the map (Fig. 3). The “make them work for it” option. If a downstream user is paying attention and realises that, for example, the side chain they are looking at is meant to be a lysine, despite the model only having atoms up to Cß, this should be the least misleading of all the options. The residue should not be mutated to, say, Alanine in this case, as that would mean you are wilfully misleading downstream users. Upon realising the atoms are missing, the downstream user can then model a (hopefully sensible) rotamer for their simulations if needed. The downside is that this approach does introduce some negative bias in favour of modelling bulk solvent into this area. Like I said, none of the options are ideal solutions.
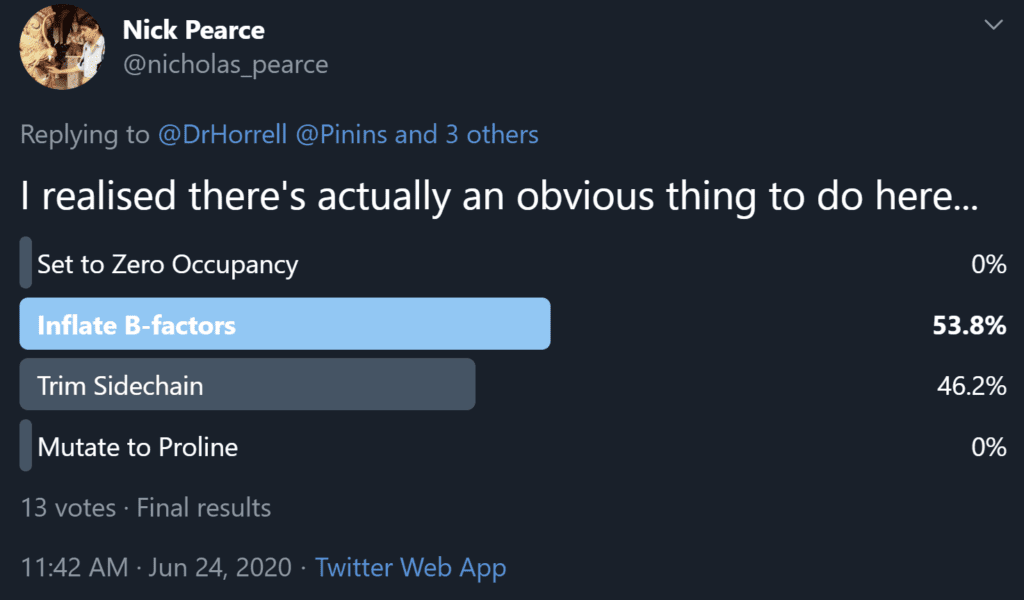
So, following this discussion between Nick Pearce, Dale Tronrud, Gianluca Santoni, Andrea Thorn, and I, we recommend option 3 as the best of the available solutions. We believe that the end goal of a crystallographic experiment should be to build atoms justified by the experimental data, i.e. the map, and leave the prediction of unobservable atoms to downstream users. We (crystallographers) are not here to “make it easier for users to avoid thinking about it”. However, after publishing the first iteration of this article a number of crystallographers made the case for option 2 on twitter and a poll of those involved resulted in 53.8% in favour of option 2 (Figure 4), so the matter is still far from resolved.
However, it’s nice to know that if we really can’t agree on the best method we can at least agree on not option 1, and there's always the fall back plan of option 4 and watch the PDB burn if we get desperate.

