Overview
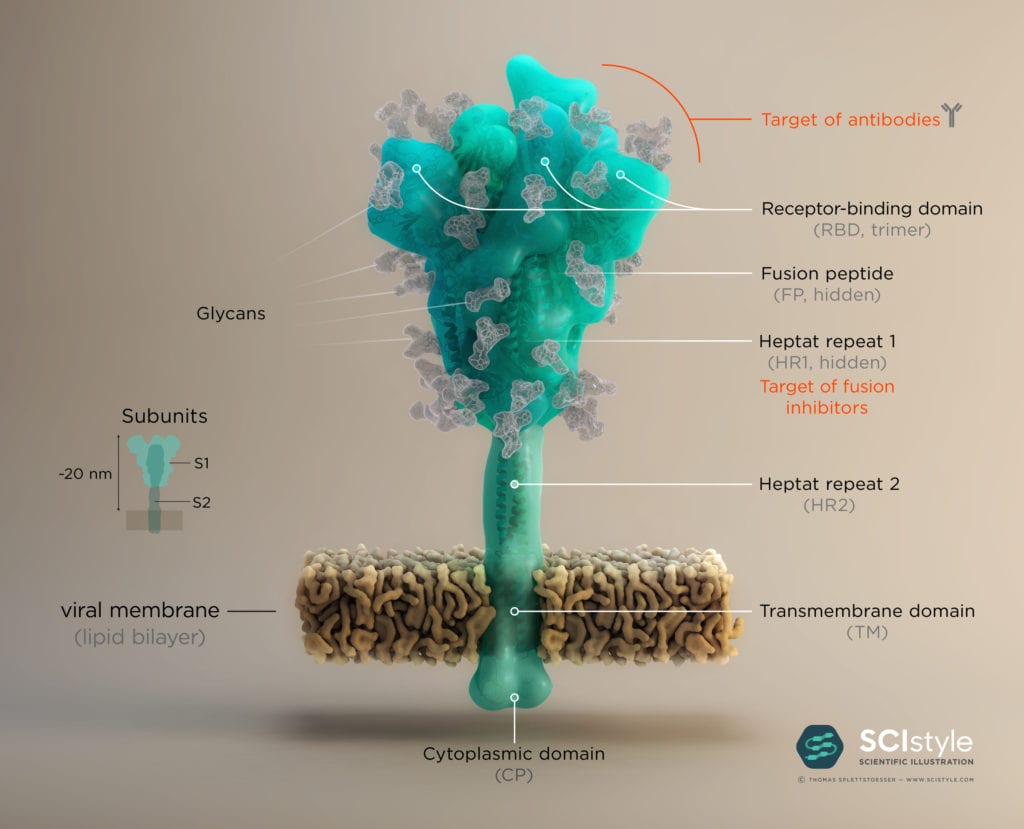
The surface proteins, also called the “spike” or S-proteins, protrude from the viral envelope of SARS-CoV-2 like “spikes of a crown”, thus giving the coronavirus its name. They mediate entry into the host cell by binding to a cellular receptor called angiotensin-converting enzyme (ACE2), triggering a cascade of events leading to membrane fusion and entry. The Spike protein is formed by three identical monomers, each consisting of the two subunits S1 and S2. Subunit S1 comprises a receptor binding domain (RBD), which interacts with ACE2 on human epithelial cells. ACE2 is a type I membrane protein expressed in lungs, heart, kidneys, and intestines, and takes part in maturation of angiotensin, a peptide hormone which controls vasoconstriction and blood pressure.
Binding Mechanism
To engage the ACE2 receptor, the RBD of S1 undergoes a hinge-like conformational rearrangement that transiently exposes the residues necessary for receptor binding. The hepta-repeat 1 and 2 domains (HR1 and HR2) play a key role in mediating fusion and entry (see Fig. 1). The exact mechanism of entry and fusion of SARS-CoV-2 with and into the host cell is still not fully established, but it is likely that the fusion mechanism is similar to SARS-CoV. The putative mechanism is that after RBD binds to the ACE2 receptor, the S2 subunit binds to the host membrane via a fusion peptide (FP), and changes conformation to trigger the association between the HR1 and HR2 domains to form the “fusion core”, which brings the viral and cellular membranes in close proximity for fusion.
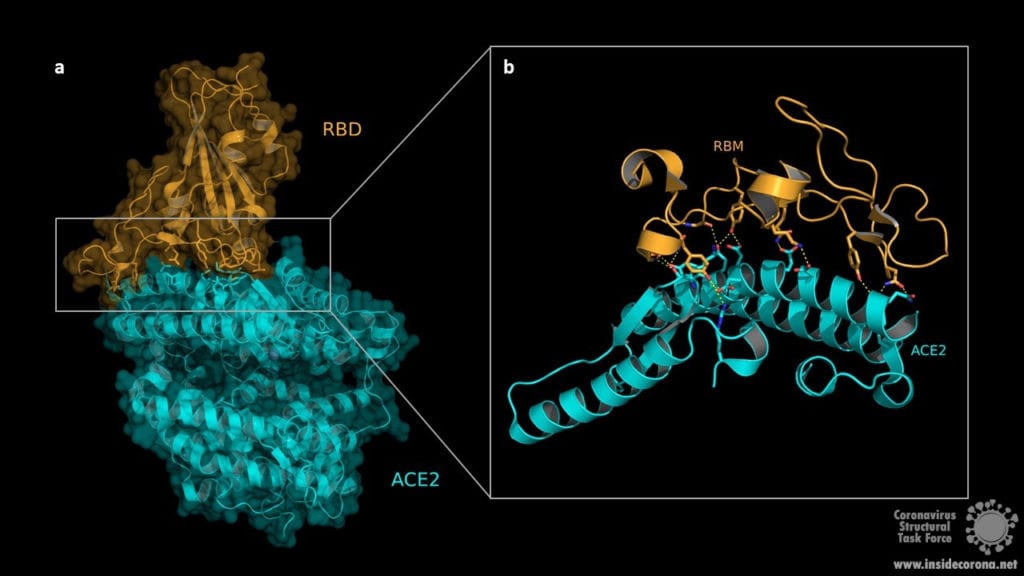
The structure of the RBD in complex with the human ACE2 receptor reveals that the interaction occurs via the spike protein RBD and the ACE2 N-terminal peptidase domain. The RBD consists of a twisted five stranded antiparallel β-sheet (β1, β2, β3, β4 und β7) forming the core together with short connecting α-helices, β-sheets and loops. These short α-helices, β-sheets and loops constitute the receptor binding motif (RBM) which is located as an extended insertion between two β-strands (β4 and β7) and contains most of the ACE2 contacting residues. The ACE2 N-terminal peptidase domain consists of two lobes that form the substrate binding site. The contact between the RBM and ACE2 is made at the bottom side of the ACE2 small lobe, with a concave outer surface in the RBM accommodating the N-terminal helix of the ACE2 and thus generating an interface of 1687Å2 (see Fig. 2).
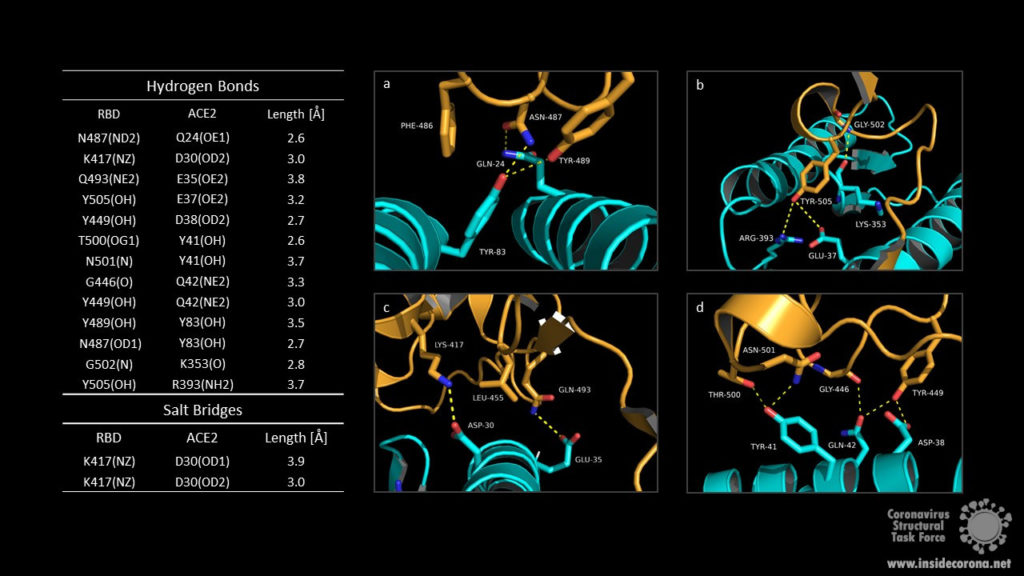
The RBM/ACE2 interface contains a network of different interactions, including hydrophilic interactions with 13 hydrogen bonds and 2 salt bridges which are shown in Fig.3. Key residues for receptor binding include the amino acids Leu-455, Phe-486, Gln-493, and Asn-501. The RBD residues Gln-493 and Asn-501 form hydrogen bonds with the respective ACE2 residues Glu-35 and Tyr 41. Phe-486 interacts with the ACE2 amino acids Gln-24, Leu-79 as well as Tyr-83 and makes contact to Met-82 by Van-der-Waals forces. Another important interaction takes place between the non-polar RBD Leu‑455 and ACE2 Asp-30, Lys-31 and His-34. Outside the RBM the amino acids Lys-417 and the ACE2 Asp-30 contribute to receptor binding by forming a salt bridge. Binding of the host cell receptor by subunit S1 destabilizes the prefusion trimer and triggers a structural rearrangement resulting in cleavage and shedding of the S1 subunit and transition of the S2 subunit to a stable postfusion conformation.
The Role of Glycosilation
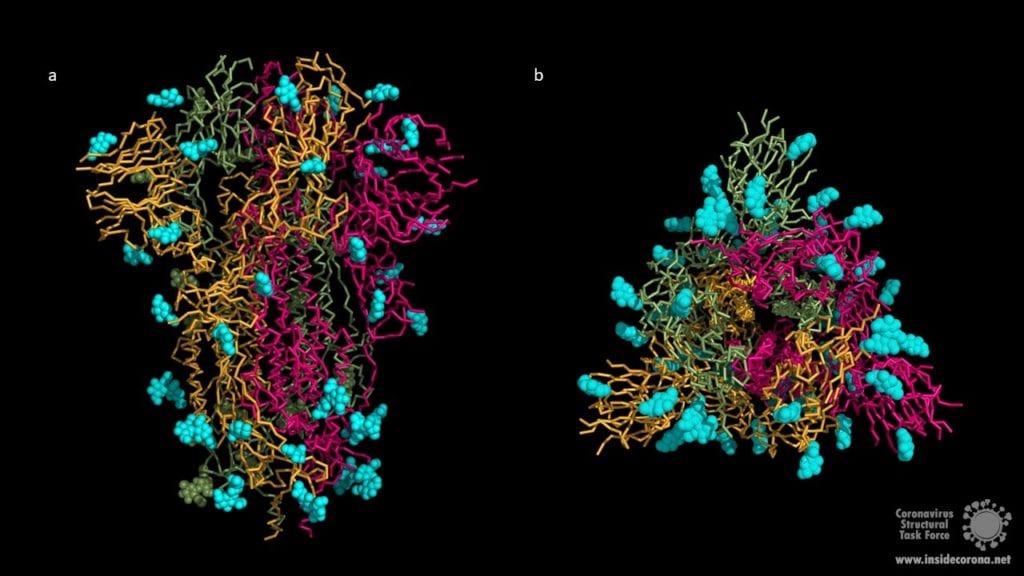
The surface of coronavirus spike proteins is densely decorated with heterogenous N-linked glycans protruding from the trimeric surface. SARS-CoV-2 spike comprises 22 N-linked glycosylation sequons per protomer. N-linked glycans play a key role in proper protein folding and in priming for fusion by host proteases. Glycans can also shield the amino acid residues and other epitopes from cells and antibody recognition, so glycosylation enables the coronavirus to evade both the innate and adaptive immune responses. It may also play a role in binding to the host cell. Unfortunately, both X-ray crystallography and cryo-EM cannot resolve long carbohydrate chains, so the structures (below) contain a maximum of three sugars. In most cases, the carbohydrate chains are much longer, covering most of the contact surfaces of the upper spike protein.
Summary
The spike protein acts as key molecule for fusion and entry, so development of drugs directly targeting this protein may be essential to contain the COVID-19 pandemic. "Capping" the spike proteins with antibodies would interrupt infection. Binding of antibodys to S1 RBD could lead to an inhibition of the RBD-ACE2 interaction, which then could prevent fusion with the host cell. In addition, in lung cells, spike functionality depends on furin-mediated pre-cleavage at the S1/S2 site for subsequent activation by TMPRSS2 (transmembrane Serinprotease 2). Thus, inhibitors of either furin or TMRPSS2 could also be considered as a potential treatment for COVID-19. As the spike protein decorates the virus hull, it could also be part of a vaccine. All of this makes the spike protein a major target in the molecular fight against COVID-19.
Further reading:
Chemistry and Engineering New: Adding the missing sugars to coronavirus protein structures
https://www.nature.com/articles/s41423-020-0426-7
https://www.nature.com/articles/s41586-020-2180-5
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7102599/
