1. Introduction:
With SARS-CoV-2 infections and related death rates continuing to rise worldwide and new variants emerging, the virus is still a great and present danger. Although we have gathered significant knowledge and the first vaccinations have started, new mutations can still set our efforts back and possibly make the virus even more potent. Thus, searches for new treatments are of paramount importance. The nucleocapsid structural protein, or N-Protein, could serve as another drug target.
The nucleocapsid’s main function is to protect the genomic RNA by packaging it into a ribonucleoprotein complex (RNP). Apart from this, the protein has other functions essential for the viral life cycle. It is involved in virion assembly, viral RNA synthesis, transcriptional regulation of genomic RNA, and translation of viral proteins1.
2. Structure
The SARS-CoV-2 nucleocapsid is an RNA-binding protein separated into five domains. Three of the domains are intrinsically disordered, meaning they are challenging for conventional structural characterization . Two of these intrinsically disordered regions (IDRs) are located at the N- and C-terminus of the protein, and the third acting as a linker between the two structured domains. Not much is known about the IDRs: their transient structural details are mostly predictions from molecular simulations2. The other two domains, the RNA-binding domain and the Dimerization domain (DMD), are well organised and their structures have been determined by X-Ray diffraction and NMR.

Disordered Domains:
The Linker
Flexible linkers contain a large number of polar and charged amino acids. The resulting electrostatic repulsion and lack of a stabilizing hydrophobic core prevents a well-structured conformation resulting in the disorder. Experiments have shown that the Linker region of SARS-CoV-2 nucleocapsid incorporates such polar regions which are repelled by the neighbouring folded domains. The Linker contains a positively charged serine and arginine rich motif which likely functions as a phosphorylation site for a direct interaction with RNA, M (membrane) protein, and Nsp31,2. Simulations reveal that the Linker does not often adopt helical conformation as they are transient, but it may contribute to oligomerization or act as a recognition motif for the binding of other proteins. Intrinsically disordered regions, in general, are thought to be involved in a number of regulatory functions including modulation of transcription, translation, post-translational modifications such as phosphorylation, and cell signalling, often through ordering when in contact with another protein domain1.
The disordered N- and C-terminal domains
The N- and the C-terminal regions of the nucleocapsid protein are also disordered but have several regions which also may form transient helices2. The N-terminal conformation is significantly affected by the neighbouring folded RNA binding domain. Electrostatic interactions with the RNA binding domain are proposed to cause a repulsion of the positively charged N-terminal domain from its positive surface of the and an attraction to the slightly negatively charged parts2.
The other disordered tail, the C-terminal domain, interacts with the neighbouring folded dimerization domain, competing with intradomain interactions2.
Folded domains:
The RNA-binding domain and dimerization domain are well-organised folded domains. They make up 257 of the 422 residues in nucleocapsid. All five domains, nevertheless, have been proposed to be involved in RNA-binding1.
The RNA-binding domain
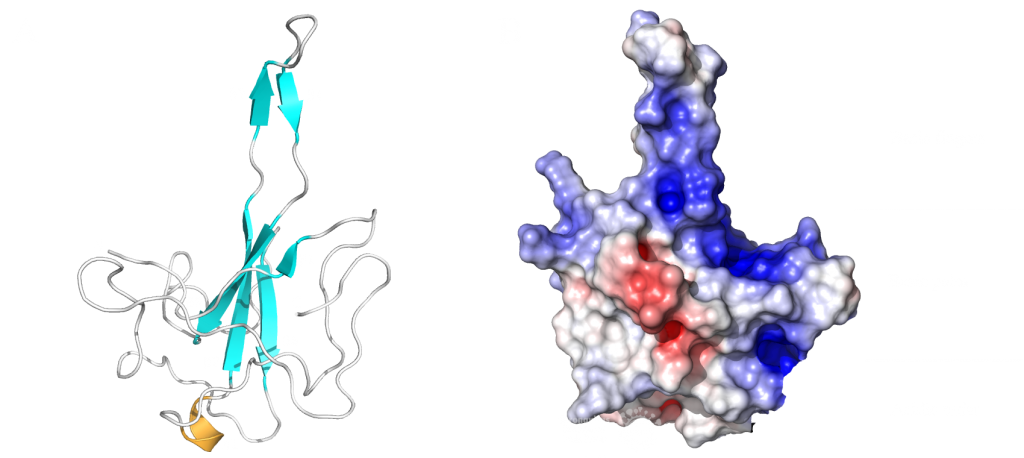
This domain mainly interacts through residues in a positively charged β-hairpin and the so-called palm region. It is rich in aromatic and basic residues that are folded into a right-hand-like shape with a protruded basic finger, a basic palm, and an acidic wrist (see Fig.1 B). Crystal structures from SARS-CoV-2 show two right-handed loops that surround the β-sheet core in a sandwiched structure. The β-sheet core consists of four antiparallel β-strands, a short 310 helix in front of the β2 strand and a protruding β hairpin that is located between the β2 and β5 strands (see Fig. 1 A). The structural basis for RNA binding by nucleocapsid is not yet known but comparisons with the less dangerous virus type HCoV-OC43 suggest a unique potential RNA binding pocket beside the β-sheet core3,4.
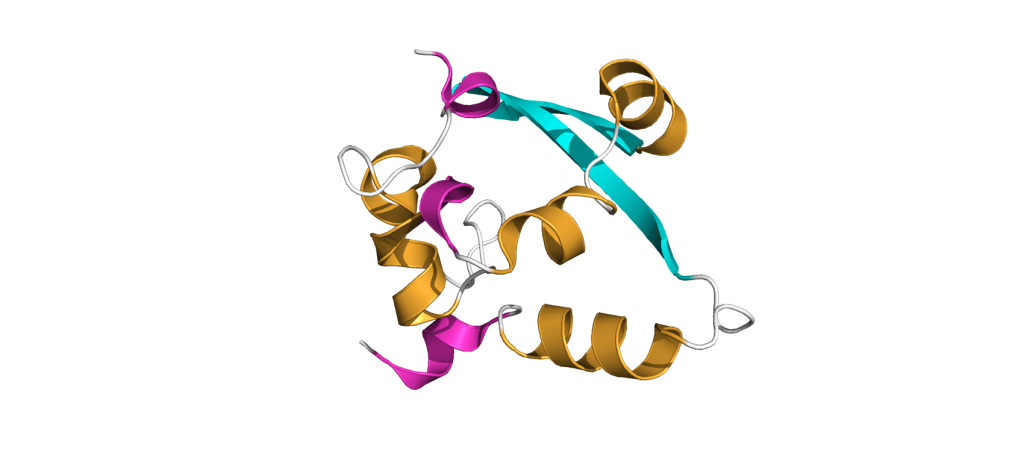
The dimerization domain (DMD)
The dimerization domain (DMD) is only stable when several nucleocapsid molecules form a dimer or oligomer. Its structure consists of three 310 -helices, five α-helices and two antiparallel β-strands, which create a β-hairpin. This β-hairpin together with the other parts of the domain form a shape that is like the letter “C”. Two domains form a tight homodimer with a rectangular slab shape, the β-hairpins from each N-Protein are at one side and the helices at the opposite side. The dimer is stabilized through hydrogen bonds and hydrophobic interactions. It is possible that the DMD has RNA binding activity, experiments showed that the amount of free RNA from SARS-CoV 2 is decreased if DMD proteins are added4,5.
The PDB currently has 22 structures that picture the RNA binding domain and the dimerization domain. The structures: 7ACT and 7ACS are particularly interesting because they are the only structures that are in complex with RNA. The RNA binding domain is also a potential inhibitor target and a subject of inhibitor Studies6. The dimerization domain has structures that show the domain as a monomer and a dimer. There is no structure of the whole protein in the PDB yet.
3. Ribonucleoprotein Complex (RNP):
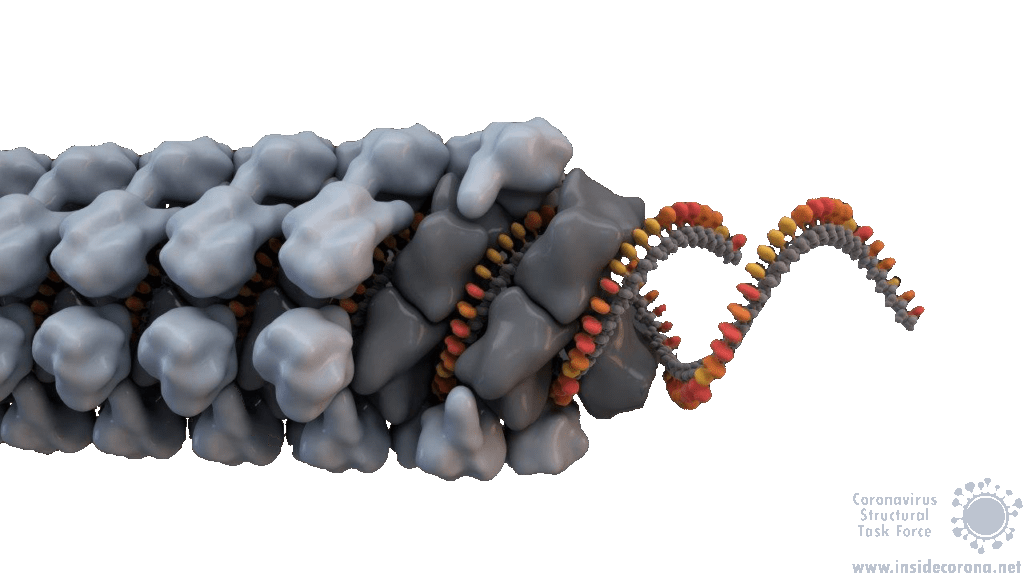
In order to package the viral RNA genome, the nucleocapsid binds the RNA via the RNA binding domain in order to form a long, flexible, helical ribonucleoprotein complex1. Two key functionalities are necessary for this process: The nucleocapsid must interact with the nucleic acid, which is preferentially mediated by GGG motifs from the leader RNA sequences7 and the nucleocapsids need the ability to oligomerize. They interact with the RNA at multiple sites through specific (sequence dependent) and non-specific (sequence independent) binding. Little is known about specific binding to the RNA, but nonspecific binding is likely to involve interactions between the negatively charged phosphate backbone of the RNA and the positively charged groove formed by the residues 248-280 of the N protein. It seems also clear that the nucleocapsid helps RNA folding1. The helical RNPs consist of coils 9 – 16 nm in diameter with a hollow interior 3 – 4 nm wide. It is frequently twisted upon itself and most of the RNPs are supercoiled into compact intertwined structures1. New cryoelectron tomography analysis of SARS-CoV-2 revealed another potential structure of the RNP, this structure is described like ‘beads on a string’ that links RNPs together, and more research is urgently needed[16]. In addition to this, the exact mechanisms of RNA protection through the nucleocapsid are still unknown1.
4. Functions:
Nucleocapsids are multifunctional proteins necessary for the viral life cycle. The main function of the nucleocapsids is the packaging of the genomic RNA into Ribonucleoprotein complexes to protect the RNA. An additional function is to enhance the stability of the entire virion through interactions with the membrane protein located in the enclosing viral membrane8. These interactions are also seen in SARS-CoV-1, where the membrane protein binds directly to the nucleocapsid via an ionic interaction1. The nucleocapsid of SARS-CoV-2 is an antagonist for interferons, suppressing the host’s defence mechanisms by preventing the synthesis of antiviral proteins9. Studies in both SARS-CoV-1 and SARS-CoV-2 have shown interactions between nucleocapsid and gRNA/sgRNA which indicate a role for the nucleocapsid in viral transcription and translation1,10. The N-Protein could also have an important role during viral assembly through interactions with envelope proteins1,11.
Many of the supplementary functions of the SARS-CoV-2 N-Proteins are still up for debate. A complete atomic structure of the RNP complex would go a long way in answering these questions, but the labile nature of the full-length N-Protein makes this a difficult task1.
5. Comparison between Coronaviruses:
The Nucleocapsid is the most conserved of the structural proteins in all coronaviruses12. This has proven useful for the development of SARS-CoV-2 Rapid Antigen Tests (for example the Roche Test). The appearance of the new English SARS-CoV-2 VUI 202012/01 variant, with changes to the spike protein, strengthens the importance of having multiple drug and test targets, particularly those that are less likely to mutate, such as the nucleocapsid13.
The high sequence analogy also allows comparison between functions within the coronavirus family, therefore a comparison between related nucleocapsids from β-Coronaviruses may shed light on these proteins’ structures and functions. The two ordered domains and the C-terminal IDR share a similar topological organization with other Coronaviruses and are involved in multiple functions in the viral life cycle. A study of the coronavirus Mouse Hepatitis Virus (MHV) analysed their nucleocapsids recruitment to Replication Transcription Complexes (RTCs) and revealed an interaction between regions on its N-terminal IDR and the serine/arginine rich region of the Linker Domain with NSP3. The interactions with NSP3 stimulate RNA replication in MHV10. Experiments have shown that the nucleocapsid from SARS-CoV-1 binds to NSP3 from MHV And interactions with NSP3 have been identified in other coronaviruses as well14,15. Thus, interactions between N-Proteins and non-structural proteins (Nsps) are proposed to have a stimulating effect for the RNA synthesis in Coronaviruses as well10.
SARS-CoV-1 and MHV both exhibit helically packed RNP complexes6 however, it is believed that SARS-CoV-2 may have a different organization16. Crystal structures of the nucleocapsid RNA binding domain from SARS-CoV-1 and SARS-CoV-2 show different crystal symmetry and packaging which could mean that SARS-CoV-2 N-Proteins have other potential contacts then SARS-CoV-14. Other possible organizations include a lattice of nucleocapsid complexes with the viral RNA linked to neighbouring RNPs like ‘beads on a string’. This ‘string‘ structure allows an efficient way of packing the large RNA genome and ensures the virus particles a high steric flexibility that is required for the incorporation into budding virions. The packaging mechanism of the SARS-CoV-2 nucleocapsid needs to be explained before we can expect to deduce an effective therapeutic approach or vaccination mechanism17.
Sources:
- 1.McBride R, van Zyl M, Fielding B. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses. Published online August 7, 2014:2991-3018. doi:10.3390/v6082991
- 2.Cubuk J, Alston JJ, Incicco JJ, et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Published online June 18, 2020. doi:10.1101/2020.06.17.158121
- 3.Peng Y, Du N, Lei Y, et al. Structures of the SARS ‐CoV‐2 nucleocapsid and their perspectives for drug design. EMBO J. Published online September 11, 2020. doi:10.15252/embj.2020105938
- 4.Kang S, Yang M, Hong Z, et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceutica Sinica B. Published online July 2020:1228-1238. doi:10.1016/j.apsb.2020.04.009
- 5.Zhou R, Zeng R, von Brunn A, Lei J. Structural characterization of the C-terminal domain of SARS-CoV-2 nucleocapsid protein. Mol Biomed. Published online August 6, 2020. doi:10.1186/s43556-020-00001-4
- 6.Chang C, Hou M-H, Chang C-F, Hsiao C-D, Huang T. The SARS coronavirus nucleocapsid protein – Forms and functions. Antiviral Research. Published online March 2014:39-50. doi:10.1016/j.antiviral.2013.12.009
- 7.Lutomski CA, El-Baba TJ, Bolla JR, Robinson CV. Proteoforms of the SARS-CoV-2 nucleocapsid protein are primed to proliferate the virus and attenuate the antibody response. Published online October 6, 2020. doi:10.1101/2020.10.06.328112
- 8.Lu S, Ye Q, Singh D, Villa E, Cleveland DW, Corbett KD. The SARS-CoV-2 Nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Published online July 31, 2020. doi:10.1101/2020.07.30.228023
- 9.Mu J, Fang Y, Yang Q, et al. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. Published online September 15, 2020. doi:10.1038/s41421-020-00208-3
- 10.Cascarina SM, Ross ED. A proposed role for the SARS‐CoV‐2 nucleocapsid protein in the formation and regulation of biomolecular condensates. FASEB j. Published online June 20, 2020:9832-9842. doi:10.1096/fj.202001351
- 11.Chen H, Cui Y, Han X, et al. Liquid–liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell Res. Published online September 8, 2020:1143-1145. doi:10.1038/s41422-020-00408-2
- 12.Chechetkin VR, Lobzin VV. Ribonucleocapsid assembly/packaging signals in the genomes of the coronaviruses SARS-CoV and SARS-CoV-2: detection, comparison and implications for therapeutic targeting. Journal of Biomolecular Structure and Dynamics. Published online September 9, 2020:1-15. doi:10.1080/07391102.2020.1815581
- 13.Diagnostics Roche. SARS-CoV-2 Rapid Antigen Test. diagnostics.roche. Published February 22, 2021. Accessed February 22, 2021. https://diagnostics.roche.com/global/en/products/params/sars-cov-2-rapid-antigen-test.html
- 14.Hurst KR, Ye R, Goebel SJ, Jayaraman P, Masters PS. An Interaction between the Nucleocapsid Protein and a Component of the Replicase-Transcriptase Complex Is Crucial for the Infectivity of Coronavirus Genomic RNA. JVI. Published online July 21, 2010:10276-10288. doi:10.1128/jvi.01287-10
- 15.Cong Y, Ulasli M, Schepers H, et al. Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle. Dutch RE, ed. J Virol. Published online November 15, 2019. doi:10.1128/jvi.01925-19
- 16.Hurst KR, Koetzner CA, Masters PS. Characterization of a Critical Interaction between the Coronavirus Nucleocapsid Protein and Nonstructural Protein 3 of the Viral Replicase-Transcriptase Complex. Journal of Virology. Published online June 12, 2013:9159-9172. doi:10.1128/jvi.01275-13
- 17.Klein S, Cortese M, Winter SL, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Published online June 23, 2020. doi:10.1101/2020.06.23.167064
