A guest entry by Hauke Hillen
In order for the novel coronavirus SARS-CoV-2 to replicate, it has to achieve two basic tasks: It needs to make copies of its genome that can be packaged into new virus particles, and it needs to activate viral genes to produce the proteins that actually form new virus particles, such as spike or nucleocapsid. Both tasks are carried out by a specialized molecular copying machine called the replication and transcription complex, or short RTC. The RTC is made up of a number of viral non-structural proteins (nsps) which act together to produce copies of the viral RNA genome. Some of the RTC components have been discussed in previous posts, for example the exonuclease nsp14, which can correct errors that occur during RNA copying (this is called proofreading),or the methyltransferases nsp14 and nsp16 that add chemical modifications to the RNA that help stabilize and hide it from the immune system (this is called capping).
In this post, we will have a closer look at the enzyme that carries out RNA replication, the RNA-dependent RNA polymerase (RdRp) nsp12, and how it interacts with the other components to form the RTC.
RNA polymera…what?
First off, let’s briefly discuss what a “RNA-dependent RNA polymerase” is, and why it is important for the virus. Polymerases are enzymes found in every living cell carrying out one of the most fundamental tasks in biology: they replicate genetic information. While most cells store their genetic information in form of DNA (deoxyribonucleic acid) and use RNA (ribonucleic acid) only as transient messenger molecules (mRNAs), some viruses rely on RNA for both information storage and transmission and are hence called “RNA viruses”. To replicate their genetic information, they need a polymerase that uses RNA as template to copy the encoded information into a new RNA molecule – a RNA-dependent RNA polymerase. Chemically speaking, RNA is a polymer (a long string of nearly-identical individual building blocks) composed of the four nucleotides adenosine (A), guanosine (G), cytosine (C) and uracil (U), the sequence of which defines the genetic information.
The job of the RNA polymerase is to read the sequence of nucleotides in a template RNA and synthesize new RNA with the same sequence (technically with a complementary sequence) using individual nucleotides as building blocks. To do this, the RNA polymerase has to achieve three basic steps. First, it needs to bind the template RNA. Second, it has to read the sequence of nucleotides in the RNA. Third, it has to incorporate the correct matching nucleotide building blocks to polymerize a new RNA strand.
Coronaviruses are RNA viruses and therefore have an RNA-dependent RNA polymerase (RdRp, or nsp12). However, they are exceptional in several ways. First, their RNA genomes are almost 30.000 nucleotides in length, which are the largest RNA virus genomes known to date. Second, their RNA polymerase nsp12 requires the additional proteins nsp7 and nsp8 in order to form the active “core” RdRp. Third, this core RdRp assembles with further viral proteins to form the RTC, which can carry out additional functions such as proofreading to remove copying errors - a highly unusual capability for RNA viruses.
Since the RNA polymerase has such a fundamental job during virus replication, it is an attractive drug target to combat viral infections. Indeed, many successful anti-viral drugs against Hepatitis C virus, HIV or Herpes virus act by inhibiting viral polymerases. Strikingly, viral RdRp enzymes are remarkably similar in their overall structure even between unrelated viruses, indicating that they share a common evolutionary ancestor and their function is so essential that it does not allow for drastic changes. Therefore, some known anti-virals developed to treat other viral diseases have also been tested for their activity against the polymerase of SARS-CoV-2 and even approved for clinical use against COVID-19 [1]. However, these repurposed compounds are generally not as effective as many had hoped, because even rather subtle structural differences between the polymerase enzymes of different viruses can have strong effects on the action of anti-viral drugs. Thus, more specific and efficient drugs against SARS-CoV-2 are therefore badly needed. In order to discover and develop such compounds, detailed knowledge of the structure and function of the RdRp is necessary.
Structure of the coronavirus RNA-dependent RNA polymerase (RdRp)
The first structure of a coronavirus polymerase was determined shortly before the outbreak of the current COVID-19 pandemic when Kirchdoerfer and Ward reported the cryo-electron microscopy (cryo-EM) structure of SARS-CoV-1 RdRp [2]. Since SARS-CoV-2 has emerged, scientists all over the world have been racing to determine the structures of its RdRp. As of February 2021, this has led to more than 20 structures of SARS-CoV-2 polymerase-complexes published in the PDB, and often several groups of scientists reported similar structures around the same time.
These structures show that the RNA polymerase nsp12 resembles a right hand with individual domains called palm, fingers and thumb (Figure 1) [3–7]. This “hand” shape is typical for viral RNA polymerases and holds a tight grip on the double-stranded RNA helix that forms between template and product strand during RNA synthesis. Within the palm lies the “active center” of the enzyme, where nucleotides are added to the growing product chain. The active center is accessible from the surface of the enzyme through a special tunnel, so that nucleotides can enter the substrate-binding site. As the template strand is opposite to the substrate-binding site, each nucleotide entering is “sampled” for whether it can form base-pairing interactions with the template base. If this is the case, the nucleotide remains bound, and it is added to the 3’ end of the product strand by forming a chemical bond. After that, the RdRp enzyme must slide ahead on the template strand by one nucleotide, which moves the newly produced 3’ end of the product RNA from the substrate-binding site (which is sometimes also referred as position +1) to the position where the previous 3’ end of the product was located prior to addition (position -1). This “translocation” completes the nucleotide addition cycle, as it positions the next templating base and frees up the substrate-binding site for the next matching building block.
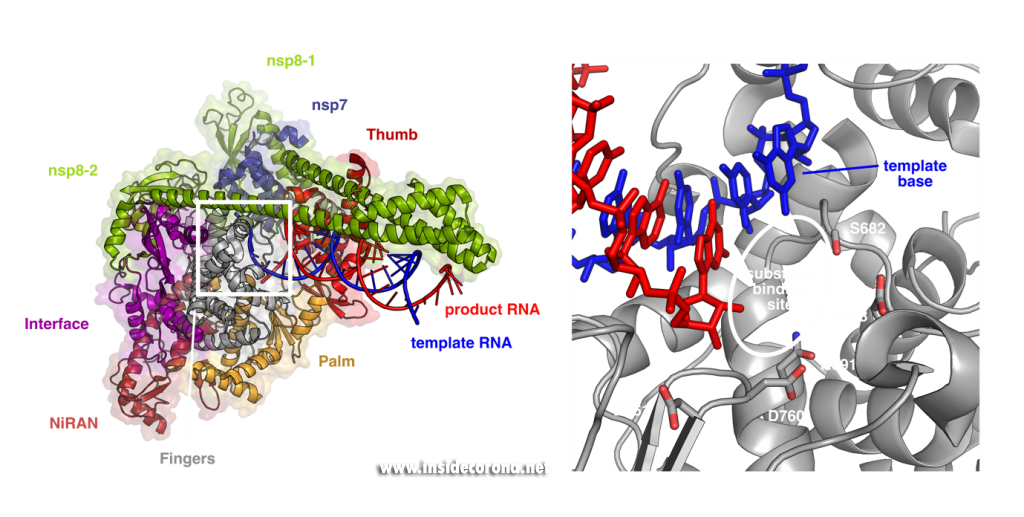
Left: Cryo-EM structure of SARS-CoV-2 RdRp (PDB 6YYT). Right: Enlarged active site with template strand in blue and nascent chain in red..
In addition to its polymerase domain, RdRp also contains a part that is only found in nidoviruses (the virus family that coronaviruses belong to) called “nidovirus RdRp-associated nucleotidyltransferase domain”, or short NiRAN-domain. Scientists believe that this domain has the capability to transfer nucleotidyl-residues, which means that it can form chemical bonds between nucleotides and other molecules. This hints that it may be involved in modification of the RNA (so-called “capping”) or in helping the enzyme initially kickstart RNA synthesis, but its precise role during coronavirus replication is still being studied by scientists.
In order to efficiently copy RNA, nsp12 requires two additional viral proteins, nsp7 and nsp8. The structures of coronavirus RdRp show that two molecules of nsp8 and one molecule of nsp7 bind on top of the hand-shaped nsp12. Interestingly, even though identical in amino acid sequence, the two nsp8 molecules adopt slightly different shapes. While one of them interacts with the finger domain of nsp12 directly, the interaction of the other one is mediated by nsp7. Both nsp8 molecules have long “arms” that protrude away from the polymerase and touch the RNA duplex as it emerges from the polymerase during replication. These “sliding poles” are unique to coronaviruses and most likely stabilize the RdRp on the RNA, which may help to make sure it doesn’t fall off during replication of the very large genome.
Seeing is believing - visualizing how anti-viral compounds block RdRp
So how exactly can this structural knowledge help to find new drugs against COVID-19? In many ways, enzymes are like tiny molecular machines. By studying their structure, one can analyze in detail how they work biochemically, and this in turn allows us to come up with ways to block their function. Most known anti-viral drugs that target RNA polymerases are so-called nucleoside analogs, which means they are molecules that structurally resemble the natural building blocks of RNA. These compounds can “trick” the RdRp by binding to the active site, but due to their chemical nature, they either cannot be incorporated into the product or lead to mutations that end the viral life cycle. The structures of SARS-CoV-2 RdRp reveal the exact architecture of the active site and show how the chemical environment that the enzyme creates around the product RNA and the substrate nucleotides, facilitates polymerization (Figure 1). This knowledge can help to rationally design or improve compounds in such a way that they bind more efficiently.
This is exemplified by recent studies analyzing how repurposed anti-virals inhibit SARS-CoV-2 RdRp. One such drug is Remdesivir, a compound originally developed against Ebola and other viruses that has also been approved by the FDA and European agencies for treatment of COVID-19 (see also this previous post). Remdesivir chemically resembles adenosine triphosphate (ATP), but has an additional bulky chemical residue called a cyano-group attached to the C1-atom of its ribose moiety. In contrast to most nucleoside analogs, Remdesivir does not block the RdRp immediately, but only after another three nucleotides are added, a process called “delayed stalling” [8–10]. Initial structures of SARS-CoV-2 RdRp in the presence of Remdesivir showed how it can act as an adenosine analog and how it can be incorporated at the 3’ end of the nascent product RNA strand and translocated to the -1 position (Figure 2a,b) [5,6]. However, this could not explain how it would lead to inhibition of RNA synthesis.
To pinpoint why Remdesivir interferes with RNA synthesis exactly after three subsequent nucleotides are added, the authors of a recent study used a combination of synthetic chemistry and structural biology [11]. They systematically determined structures of SARS-CoV-2 RdRp bound to a template-product RNA duplex which contained Remdesivir and either two or three additional nucleotides at the 3’ end. In the first case, the structure showed that the RdRp was in the post-translocated state, with Remdesivir at position -3 and an empty substrate binding site, as expected (Figure 2c). In contrast, the structure of the RdRp with an RNA containing Remdesivir and three additional nucleotides was not in the post-translocated state and Remdesivir was not located at position -4. Instead, it remained at position -3, and the third additional nucleotide at the 3’ end of the product was stuck in the substrate-binding site (Figure 2d). This state resembles the situation directly after addition of a new nucleotide but before translocation and is hence called the pre-translocated state. This suggests that remdesivir inhibits the SARS-CoV-2 RNA polymerase by posing a translocation barrier, and the structures provide a molecular and chemical explanation for this: Initially, Remdesivir can be added to the growing RNA just like adenosine triphosphate and also translocated to add another two nucleotides. However, after binding and addition of a third nucleotide, Remdesivir can not be translocated to the -4 position, because its bulky cyano group would clash with a serine residue (Ser861) in the thumb domain of nsp12 (Figure 2c). Therefore, the polymerase gets stuck in the pre-translocated state, which explains why exactly three nucleotides can be added after Remdesivir incorporation – addition of a fourth nucleotide would first require translocation. This proposed mechanism is in agreement with previous modelling [5,10] and was shortly after independently confirmed by another structural study, in which the authors managed to trap an identical pre-translocated, stalled intermediate with Remdesivir in position -3 [12].
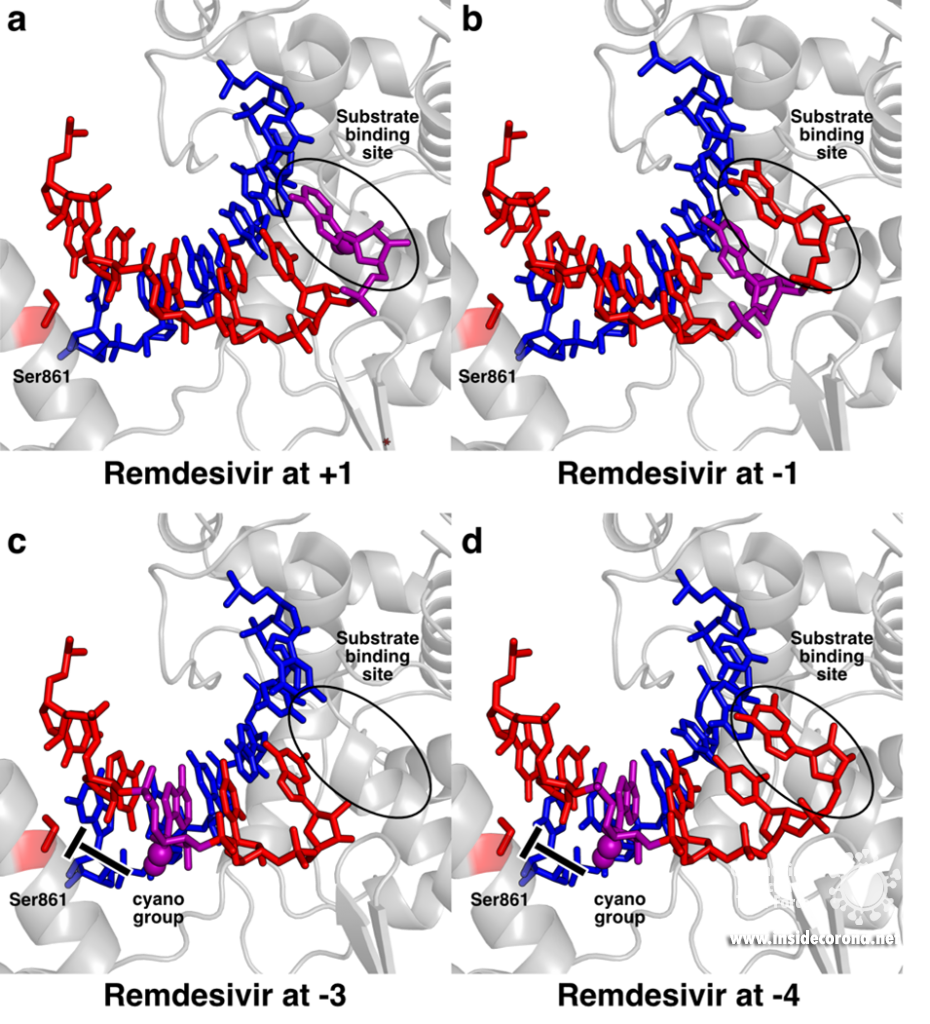
Structural snapshots of remdesivir (purple) moving through the active site of SARS-CoV-2 RdRp. When it reaches the third position after its incorporation to the RNA (-3), its further movement is blocked because the cyano group would bump into Ser861. A) Remdesivir at position +1 (PDB 7BV2) B) Remdesivir at position -1 PDB 7C2K C) Remdesivir at position -3 (PDB 7B3B) D) Remdesivir at position -3 with the nucleotide at the 3’ end stuck in the substrate binding site (PDB 7B3C).
This mechanism also suggests how Remdesivir may at least partially escape the coronavirus proofreading enzyme nsp14, which removes misincorporated nucleotides at the 3’ end of the RNA and thus counteracts anti-virals that target RdRp. Since Remdesivir can be translocated until it reaches position -3 before it causes stalling, it may leave the active site of the enzyme before it can be recognized by the proofreading machinery.
Importantly, these studies also provide clues as to why Remdesivir has had limited success in fighting COVID-19. The structures show that the steric block between Ser861 and the cyano group of Remdesivir is not severe and can therefore be overcome by the enzyme, for example at high concentrations of substrate NTPs [8] Consistent with this, substitution of Ser861 with residues that clash even less (Alanine or Glycine) make the RdRp less sensitive or even resistant to Remdesivir [5,13]. This suggests that the translocation barrier could potentially be enhanced by a compound that leads to more severe clashes. One way to achieve this could be to modify Remdesivir to contain more bulky chemical moieties than the cyano group. Thus, the detailed molecular insights into the mechanism of Remdesivir also provide a rational basis for designing more potent anti-virals and test their effect on the SARS-CoV-2 RdRp.
Similar structure-function studies are now being undertaken also for other promising anti-viral compounds, such as Favipiravir. Like Remdesivir, it is a nucleoside analogue that was initially developed against other viruses, but showed some promising results against SARS-CoV-2. Structures of the SARS-CoV-2 RNA polymerase with Favipiravir show how it mimics both guanosine or adenosine in the active site of the enzyme by forming unusual base-pairing interactions with cytosine and uracil, respectively, and this leads to errors during RNA copying that eventually kill the virus [14,15]. Another study recently reported the structure of Suramin bound to SARS-CoV-2 RdRp [16]. In contrast to Remdesivir and Favipiravir, this compound is not a nucleoside analog and hence does not get incorporated into the RNA. Instead, two Suramin molecules can apparently bind to the RdRp and thereby prevent its association with template and product RNA, rendering it inactive.
These studies are good examples of how structural biology can visualize complicated chemical reactions in an intuitive way. Based on these results, drugs like Remdesivir or Favipiravir can be rationally optimized to more effectively combat COVID-19 and may cause less side effects.
Dissecting the SARS-CoV-2 RTC structure by structure
In addition to provide detailed snapshots of how anti-viral compounds act to inhibit SARS-CoV-2 RdRp, structural studies are also helping scientists to understand how the unique coronavirus RTC combines different functions such as RNA synthesis, proofreading and modification. After the structure of the “core” SARS-CoV-2 RdRp was determined within a few months after the outbreak of COVID-19, scientists quickly moved to studying how additional non-structural proteins bind to it to form the RTC. One of these is nsp13, which belongs to a protein class called “helicases”. These are enzymes that bind to DNA or RNA and, with the help of chemical energy in the form of ATP, move along them or unwind helices. Cryo-EM structures of nsp13 bound to the SARS-CoV-2 RdRp show that two molecules of nsp13 can bind to the RdRp, and suggest that it may allow the polymerase to move backwards on the RNA (Figure 3) [17,18]. This finding seems unexpected at first, but experts think this may be required for the proofreading enzyme nsp14 to remove errors that the polymerase makes during copying or to produce the mRNAs for certain viral proteins. Another recent cryo-EM structure shows that the small protein nsp9 interacts with the NiRAN domain in the SARS-CoV-2 polymerase [19]. In the accompanying paper, the authors suggest that the NiRAN domain may be involved in RNA capping, and nsp9 seems to block its activity. However, others have proposed that nsp9 is in fact a substrate of nucleotidylation by the NiRAN domain, and that it may be involved in priming the RNA polymerase for initial RNA synthesis [20]. Thus, further studies are necessary to determine whether the NiRAN is involved in capping, priming or even both.
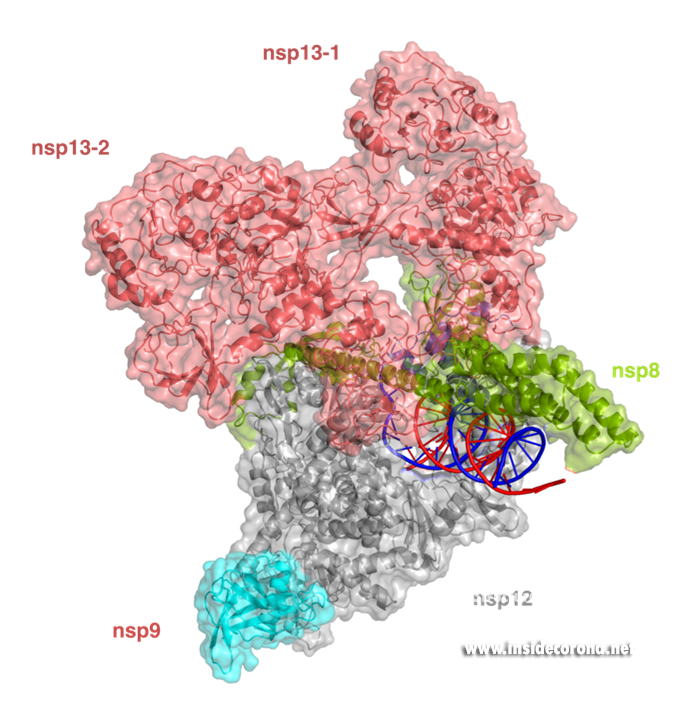
Structure of the SARS-CoV-2 RdRp complex with nsp13 (salmon) and nsp9 (cyan) (PDB 7CYQ).
What’s next?
Over the past year, scientists have uncovered the structure of the SARS-CoV-2 RdRp and associated proteins at a record-breaking pace. While these structures provide impressive first glimpses at RdRp-complexes, researchers are already working to determine how the remaining nsp proteins interact with the RdRp to form the complete RTC complex. This may ultimately aid the quest to find new treatment options for COVID-19 that not only target the polymerase itself, but also proofreading or RNA capping. Such drugs are not only desperately needed for the current pandemic, but may also prove useful for future emerging coronaviruses, because the RNA polymerase is typically very similar even between different virus strains.
1. Ledford H: Hopes rise for coronavirus drug remdesivir. Nature 2020, doi:10.1038/d41586-020-01295-8.
2. Kirchdoerfer RN, Ward AB: Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 2019, 10:2342.
3. Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, et al.: Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368:779–782.
4. Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, Cramer P: Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584:154–156.
5. Wang Q, Wu J, Wang H, Gao Y, Liu Q, Mu A, Ji W, Yan L, Zhu Y, Zhu C, et al.: Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell 2020, 182:417-428.e13.
6. Yin W, Mao C, Luan X, Shen D-D, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M, et al.: Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368:1499–1504.
7. Peng Q, Peng R, Yuan B, Zhao J, Wang M, Wang X, Wang Q, Sun Y, Fan Z, Qi J, et al.: Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2. Cell Reports 2020, 31:107774.
8. Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, Götte M: Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem 2020, 295:6785–6797.
9. Tchesnokov EP, Feng JY, Porter DP, Götte M: Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses 2019, 11:326.
10. Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Gotte M: The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. The Journal of biological chemistry 2020, doi:10.1074/jbc.ac120.013056.
11. Kokic G, Hillen HS, Tegunov D, Dienemann C, Seitz F, Schmitzova J, Farnung L, Siewert A, Höbartner C, Cramer P: Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat Commun 2021, 12:279.
12. Bravo JPK, Dangerfield TL, Taylor DW, Johnson KA: Remdesivir is a delayed translocation inhibitor of SARS CoV-2 replication in vitro. Biorxiv 2020, doi:10.1101/2020.12.14.422718.
13. Tchesnokov EP, Gordon CJ, Woolner E, Kocinkova D, Perry JK, Feng JY, Porter DP, Götte M: Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J Biol Chem 2020, 295:16156–16165.
14. Naydenova K, Muir KW, Wu L-F, Zhang Z, Coscia F, Peet MJ, Castro-Hartmann P, Qian P, Sader K, Dent K, et al.: Structural basis for the inhibition of the SARS-CoV-2 RNA-dependent RNA polymerase by favipiravir-RTP. Biorxiv 2020, doi:10.1101/2020.10.21.347690.
15. Peng Q, Peng R, Yuan B, Wang M, Zhao J, Fu L, Qi J, Shi Y: Structural basis of SARS-CoV-2 polymerase inhibition by Favipiravir. Innovation 2021, doi:10.1016/j.xinn.2021.100080.
16. Yin W, Luan X, Li Z, Zhou Z, Wang Q, Gao M, Wang X, Zhou F, Shi J, You E, et al.: Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin. Nat Struct Mol Biol 2021, 28:319–325.
17. Chen J, Malone B, Llewellyn E, Grasso M, Shelton PMM, Olinares PDB, Maruthi K, Eng ET, Vatandaslar H, Chait BT, et al.: Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell 2020, doi:10.1016/j.cell.2020.07.033.
18. Yan L, Zhang Y, Ge J, Zheng L, Gao Y, Wang T, Jia Z, Wang H, Huang Y, Li M, et al.: Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat Commun 2020, 11:5874.
19. Yan L, Ge J, Zheng L, Zhang Y, Gao Y, Wang T, Huang Y, Yang Y, Gao S, Li M, et al.: Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis. Cell 2021, 184:184-193.e10.
20. Slanina H, Madhugiri R, Bylapudi G, Schultheiß K, Karl N, Gulyaeva A, Gorbalenya AE, Linne U, Ziebuhr J: Coronavirus replication–transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc National Acad Sci 2021, 118:e2022310118.
