Due to a new outbreak of pulmonary diseases caused by SARS-CoV-2, the development of new drugs is essential to contain the COVID-19 pandemic. One promising drug target is the 3C-like protease, also known as main protease or MPro. Most of the virus proteins are translated as one long polypeptide chain, which then has to be cleaved into functional proteins. For the viral polyproteins ppa1a and ppa1ab, 11 sites at the C-terminal end (downwards from nsp4) are cleaved by main protease. As the RNA polymerase complex is part of this chain (nsp7, nsp8, nsp12 and nsp14), inhibition of this main protease stops replication.
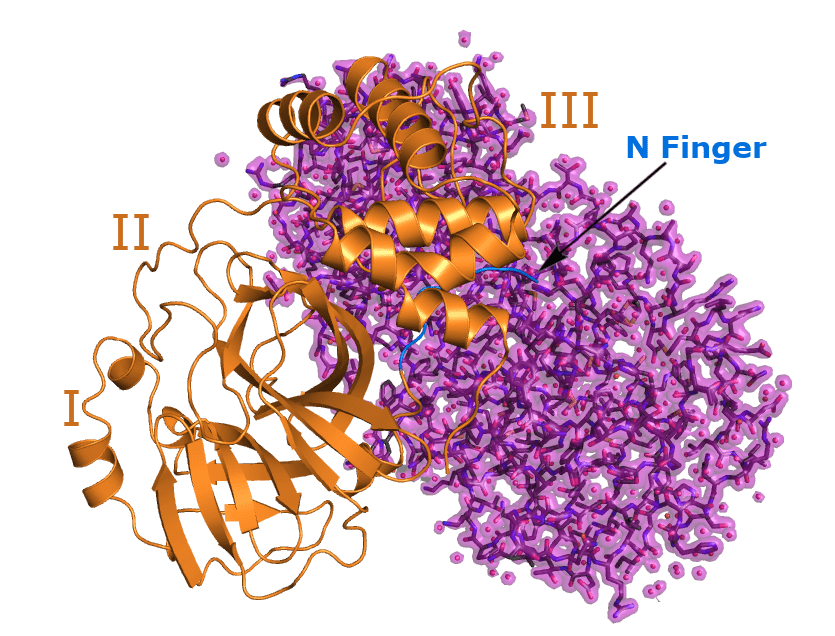
The 3C-like protease is a cysteine protease which is characterized by a catalytic dyad consisting of the amino acids cysteine and histidine. The homodimer is comprised of two perpendicular protomers forming a catalytic cleft in between. Each of these protomer is composed of three domains. Domain I and II (N-terminal domain) form an antiparallel chymotrypsin-like β-barrel structure in which the substrate binding site is located. Domain III (C-terminal end) consist of five α-helices arranged in a cluster regulating dimerization through a salt-bridge interaction between Glu‑290 of one protomer and Arg-4 of the other.
The N-terminal residues called “N-finger” (see image) make contact predominantly to domain II of the other protomer generating a contact interface of ~1394 Å2. The dimerization in essential for protease activity as the N-terminal residue Ser-1 of one protomer interacts with Glu-166 of the other protomer keeping a substrate binding site in the right shape. This substrate binding site contains a catalytic dyad consisting of the residues Cys-145 and His-41. Next to the catalytic dyad is the substrate binding pocket called S1. It consists of the side chains Phe-140, His-163 and the main chain atoms of Glu-166, Asn-142, Gly-143 and His-172. This pocket mediates the high specificity for a Gln [Leu-Gln↓(Ser,Ala,Gly)] of the substrate to be cut, as the carbonyl oxygen of this Gln is stabilized by the amino acids Gly‑143 and Cys-145.
The specific substrate binding site S1 is thought to bind an inhibitor to work as a drug. This would then inhibit the cleavage of polyproteins and hence stop replication of the virus RNA. An advantage of 3C-like protease as a drug target is that up to date, no human proteases with similar cleavage specificity are known, and as a consequence, newly designed drugs are unlikely to be toxic. The potential inhibitors can be divided into two classes based on their chemical structures: The first class involves peptide chains that fit the catalytic site of the enzyme by making a covalent link with Cys-145, therefore blocking substrate binding. The second class consists of small organic compounds that bind to the enzyme active site, acting as a competitive inhibitor and hinder the substrate from entering the active site cavity. A potential drug which belongs to the second class is Lopinavir, a HIV1 protease inhibitor, which seems to be a promising candidate for the treatment of coronavirus infections. If the efficacy of Lopinavir against SARS-CoV-19 is confirmed, it would have the advantage that it is already approved as an HIV drug for humans.
Learn more:
https://www.the-scientist.com/image-of-the-day/image-of-the-day-coronavirus-in-3d-67315
https://science.sciencemag.org/content/early/2020/03/20/science.abb3405
