The world holds its breath as the novel Coronavirus continues to spread across the world, bringing our lives to a halt. We have gathered a lot of knowledge about the virus but there are still many gaps to fill. The non-structural-protein 3 (nsp3) represents one of these gaps in our knowledge. As the largest protein encoded by the coronaviruses genome, untangling its structure and function poses a huge task.
However, we can glean some knowledge around the specific function of SARS-CoV-2 nsp3 by looking at the virus‘s subfamily, Orthocoronaviridae. As related viruses do share some common traits, academics were not completely unprepared when SARS-CoV-2 came. In the background, while only very few people were worried about a new corona virus, scientists around the world had been investigating the invisible enemy for decades. Building on this past work we look at the functions of proteins from other coronaviruse, like Murine Hepatitis Virus (MHV) and SARS-CoV, to learn more about how best to fight against SARS-CoV-2.
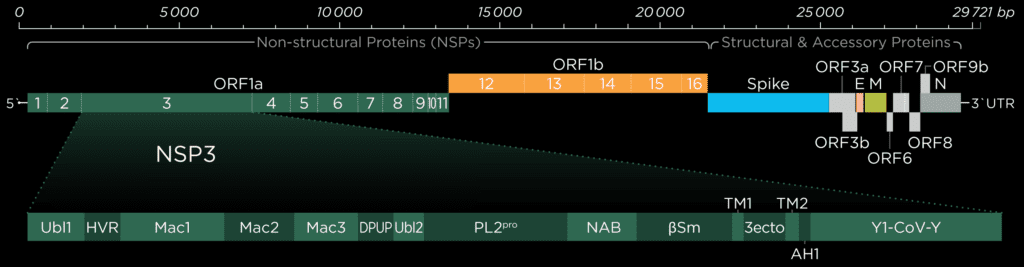
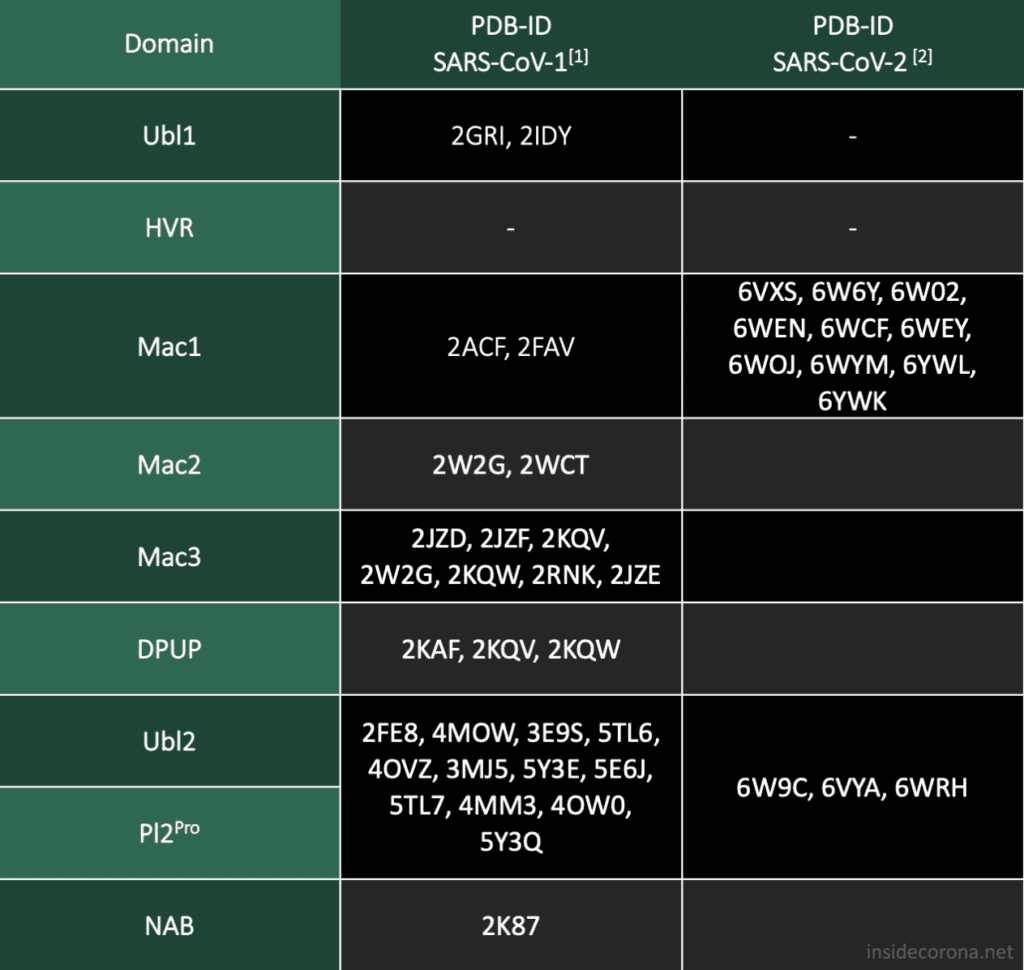
The gene which produces nsp3 lies on the open reading frame 1a (ORF1a) which encodes polyprotein 1a. The sequence for nsp3 of SARS-CoV is 1922 amino acids long and sandwiched between nsp2 and nsp4. It not only cleaves itself from the polyprotein by its papain-like protease domain but also nsp1 and nsp2. In coronaviruses, 18 different domains have been found in nsp3. Each virus type has 10 to 16 of these, out of which eight domains and two transmembrane regions form the conserved part of nsp3, which can be found in every coronavirus known to date [1]:
- Ubiquitin-like-domian (Ubl1)
- Ubiquitin-like-domain (Ubl2)
- Papain-like protease (PlPro)
- Macro domain / X domain (Mac)
- Hypervariable region / Glu-rich acidic domain (HVR)
- Transmembrane regions (TM1)
- Transmembrane regions (TM2)
- Ectodomain / Zinc finger domain (3ecto)
- Nidovirus-conserved domain of unknown function (Y1)
- Coronvirus specific carboxyl-terminal domain (CoV-Y)
To start our investigation on SARS-CoV-2 related structural data, we will look into the protein sequences of SARS-CoV and SARS-CoV-2 to learn where they are similar and where they differ.
Genetic Comparsion of SARS-CoV and SARS-CoV-2
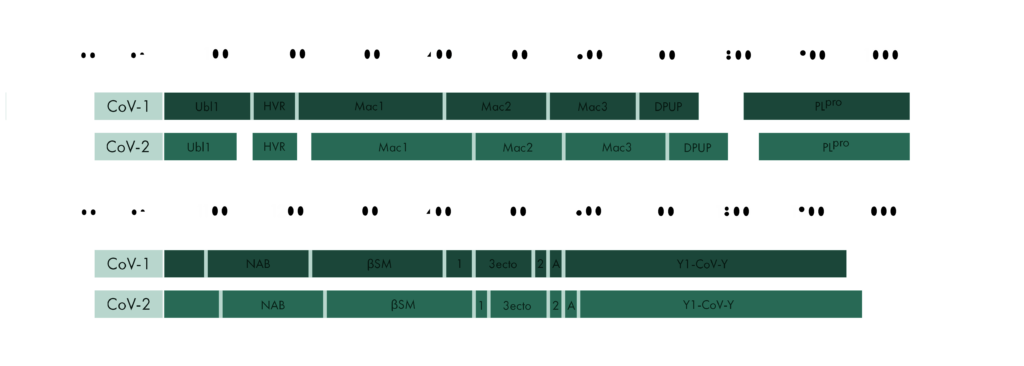
SARS-CoV has 16 domains which span 1922 amino acids. The nsp3 protein of SARS-CoV-2 is a bit longer at 1945 amino acids. When compared to each other, there is an overall similarity of 75,97%.[2] In Addition to the ten conserved domains the nsp3 gene of SARS-CoV-2 codes for four domains:
- Nucleic-acidic-binding domain (NAB)
- Betacoronavirus specific marker domain (βSM)
- Domain preceding Ubl2 and PL2pro (DPUP)
- Amphipathic helix 1 (AH1)
The two domains at the N-terminal end, Ubl1 and HVR, have an alignment of 79% and 64%, respectively. There seems to be a trend in coronaviridae for these domains to be poorly conserved, but Ubl1 still adopts the expected conserved fold.[4] If this proves true, could be analysed by comparing the sequence alignment and the structural similarity. It is unsurprising that the "high variable region" lives up to its name and shows the worst alignment of all. In the related MHV nsp3, this domain is dispensable for replication.[5]
It has been speculated that the Mac1 domain functions as an ADP ribose 1"-phosphatase, however, the effects of mutation in this region differ from virus to virus.[4] As a result, it is difficult to judge what significance the bad alignment of this domain will have on our understanding of SARS-CoV-2 without further research.
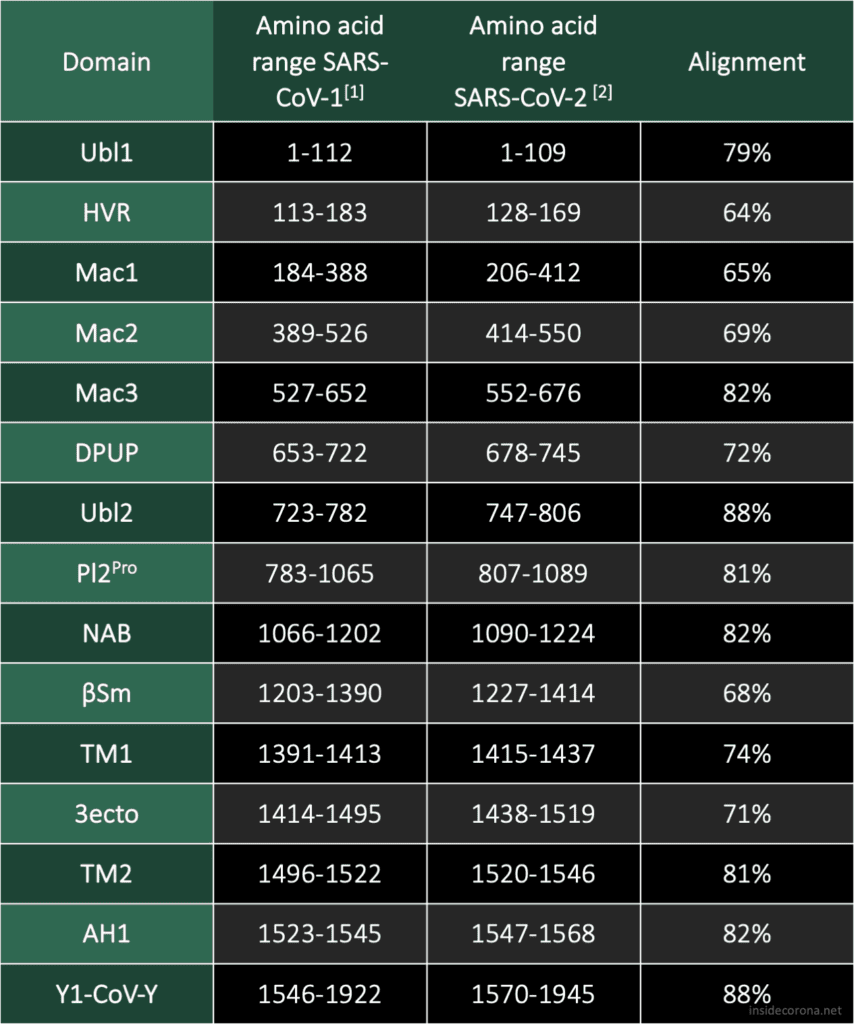
The Mac1 domain, also known as the X-domain, is followed by two macrodomains which were originally called "SARS-CoV Unique domains" (SUD-N and SUD-M), but were renamed when they were found to not be unique to SARS-CoV. It has since been observed that only Mac3 plays an essential role in viral RNA replication[6], which could explain why Mac3 is one the most conserved domains in the alignment of SARS-CoV and SARS-CoV-2.
Pl2Pro and its neighbouring domain Ubl2 show some of the highest sequence alignments of all domain comaprisons. This could be explained by their essential function to cleave nsp3 from the polyprotein.
Little is known about the domains following Pl2Pro and our current structural knowledge is limited to a nuclear magnetic resonance (NMR) structure of NAB. While the structure and function of Y1 and CoV-Y from SARS-CoV-2 are currently unknown, their sequence, which compromises a fifth of the genome, is highly conserved in all coronaviruses.
In the second part of the series of Untangling Nsp3 of SARS-CoV-2 we will delve deeper into some structures of nsp3 of SARS-CoV-1 and SARS-CoV-2 and will try to find out how the differences in the sequence may have influenced some structures of the protein. For a further in-depth reading on the topics discussed here I highly recommend the sources below.
Picture by Kristopher Nolte
Sources
- [1] Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018 Jan;149:58-74. doi: 10.1016/j.antiviral.2017.11.001. Epub 2017 Nov 8. PMID: 29128390; PMCID: PMC7113668.
- [2] Madden T. The BLAST Sequence Analysis Tool. 2002 Oct 9 [Updated 2003 Aug 13]. In: McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2002-. Chapter 16. Available from: http://www.ncbi.nlm.nih.gov/books/NBK21097/
- [3] https://github.com/thorn-lab/coronavirus_structural_task_force
- [4] Benjamin W. Neuman, Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles, Antiviral Research, Volume 135, 2016, Pages 97-107, ISSN 0166-3542, https://doi.org/10.1016/j.antiviral.2016.10.005.
- [5] K.R. Hurst, C.A. Koetzner, P.S. Masters, Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex J. Virol., 87 (2013), pp. 9159-9172
- [6] Kusov Y, Tan J, Alvarez E, Enjuanes L, Hilgenfeld R. A G-quadruplex-binding macrodomain within the "SARS-unique domain" is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015 Oct;484:313-22. doi: 10.1016/j.virol.2015.06.016. Epub 2015 Jul 3. PMID: 26149721; PMCID: PMC4567502.
