Knowing a protein’s 3D structure enables scientists to investigate its general shape and stability, deduce its potential function, and run drug-binding simulations to find a cure if the protein is from a pathogen. However, solving a structure is not an easy task, as proteins are too small to observe with optical microscopes. Furthermore, the process of collecting and preparing a protein sample and getting the final structure out of the experimental data can take several weeks or even months and is, hence, quite expensive.
In 2021, everything changed when AlphaFold2 was published: a new protein structure prediction software, which is not only incredibly accurate but also easy to use. Immediately, a great hype dominated the news, but did AlphaFold really change structural biology forever and will it make conventional methods obsolete?
Making Protein Structures Visible
Before we dive into the solution to an old problem of structural biology, we have to cover a few basics first. So, what are proteins again? In a nutshell, proteins are tiny nano-sized machines that perform any kind of task in your body. Some proteins organize the replication of your cells, while others digest your food. Another group of proteins makes up your hair, and plenty of other things in your body are also performed by proteins. A protein’s capabilities are determined by its shape, which raises great interest in its exact 3D structure. Unfortunately, proteins are smaller than visible light, so we cannot observe them with any optical microscope. Wait, smaller than light? How is this possible, you may ask? This picture should help to understand this:
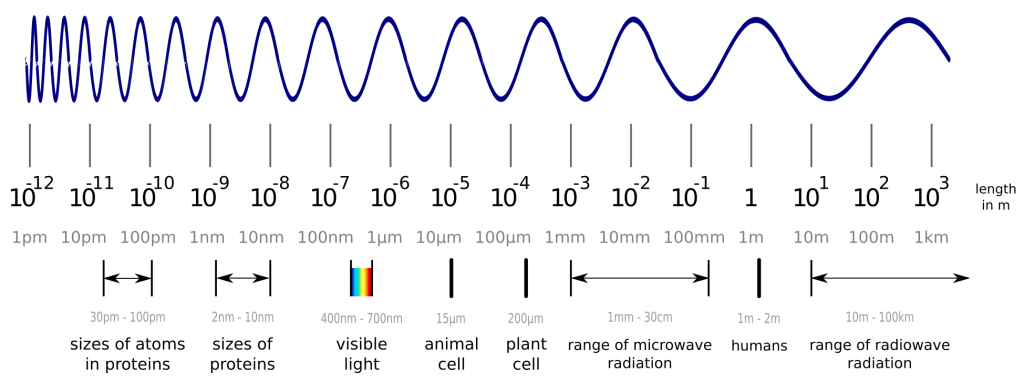
While visible light can easily interact with objects, which are larger than the wavelength of visible light itself, it does not interact much with smaller objects like proteins and atoms. Luckily, we can also create “light” in the invisible spectrum and measure the proteins with a detector instead of our eyes. One method to do this is X-ray crystallography. The details are complicated, but the important steps are to produce and purify your protein from bacteria, grow crystals out of your protein (yes, you can actually do this!) and shoot X-ray beams at it. With the data from the detector, you are able to create a 3D model of your protein, which may not represent reality perfectly, but is accurate enough to work with. You can find out more about these 3D models in another blogpost.
However, a lot can go wrong with this: The protein could kill the bacteria which should produce it for you, crystals might not form properly, and the process of turning the collected data into a model is not straightforward as well. All in all, this method can produce the desired result, but with a high investment of both, money and time.
The Protein Folding Problem
Proteins consist of long chains of amino acids. Most life on earth, us included, uses twenty different amino acids, each one with its own unique properties. For example, some are positively charged and like to be surrounded by negatively charged molecules or water. Others are not charged at all and prefer to hide inside a protein core and stay away from the water in which the protein is located. A protein’s amino acids are chained together in sequence and their properties define the shape, the final fold, of a protein. So, if you changed one amino acid in the chain into another one with vastly different properties, the whole protein would fold a bit differently. In fact, small changes might only modify the fold slightly, but multiple and huge differences in the sequence result usually in large changes in the fold. Since shape determines a protein’s function, the new protein might also do different things. The important takeaway here is: The information about the 3D structure is hidden in the sequence of amino acids. However, it remained a mystery how the folding into the final shape works in all details and could therefore not be replicated in simulations. This mystery is today known as the protein folding problem.
The hidden information in the amino acid sequence motivated many scientists around the globe to work on protein structure prediction software. Soon, those scientists started to compete against each other in the CASP competition – the Critical Assessment of Techniques for Protein Structure Prediction. Every two years since 1994, the state-of-the-art techniques have been measured against each other, but for decades all methods were not reliable enough and made many mistakes. Only recently, at CASP13 in 2018, Deepmind’s AlphaFold succeeded the first time with predictions of phenomenal quality for many of the given input sequences. Nevertheless, there was still much room for improvement. In 2020, AlphaFold2 followed and produced for the very first time in history structures almost as good as the conventionally solved ones. The protein folding problem itself has been known for more than 50 years now, but only with current technology and new approaches like the use of deep learning, structure prediction became finally real.
How accurate is AlphaFold?
When people talk about AlphaFold, they usually mean AlphaFold2. While scientists use calculations to measure the difference between a prediction and the experimentally solved structure it should resemble, the probably best way to demonstrate its accuracy to a newcomer in the field is a picture:
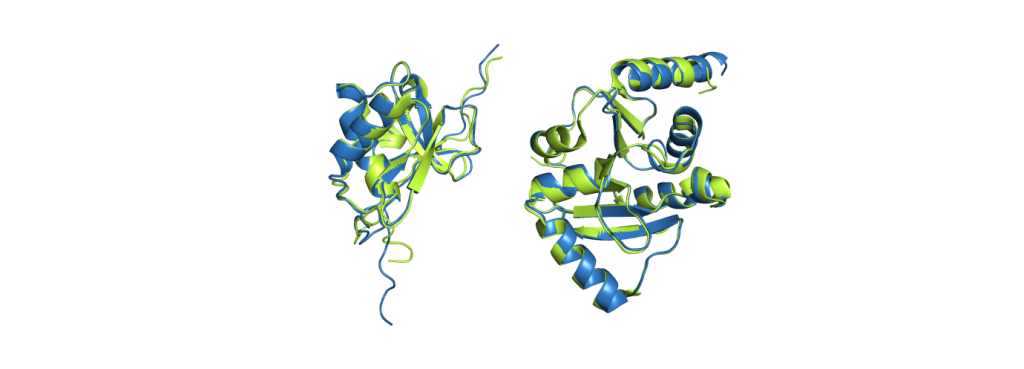
CoV-2 protein nsp3 (PDB code 7LGO), on the right is the Mac1 domain from the same protein (PDB code 6WEY).
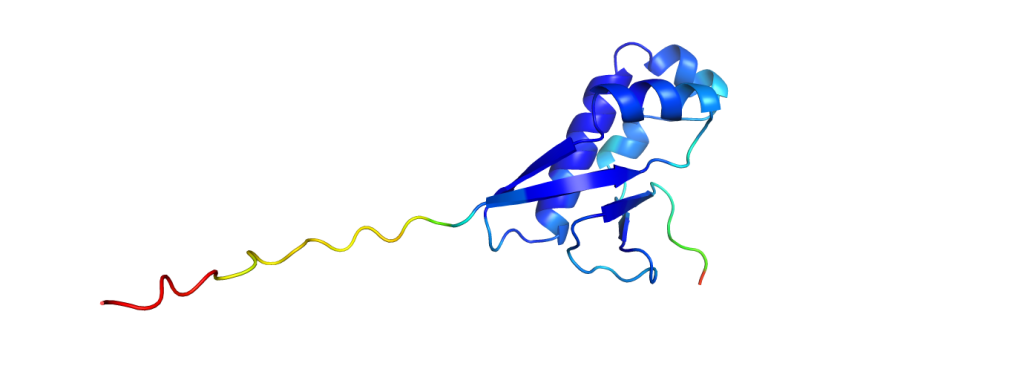
The green structures are the ones obtained from experimental data; the blue structures are predicted by AlphaFold2. It is already a challenging task to predict which parts fold into helices and sheets (the spirals and the arrows), but AlphaFold2 even predicted a correct alignment of those and also the parts in between, resulting in nearly perfect agreement. The biggest deviations between experimental structure and prediction are usually found at the ends of the chains. While it does not give such excellent results for all proteins, it still performs pretty reliably and even provides some feedback on its confidence, making it easy to spot regions with a wrong fold.
And to put the breakthrough of AlphaFold2 into perspective: to measure how similar the predictions are to experimentally solved structures, one could calculate a similarity value with one of many available metrics. One metric used in the CASP competition is the GDT, the global distance test, which returns values from 0% (no similarity at all) to 100% (identical structures). While all methods in the past did not pass on average the 60% mark, AlphaFold2 scored consistently with GDTs of over 90%.
Does AlphaFold replace conventional methods?
As we have seen, AlphaFold ist almost as accurate as the models obtained from X-ray crystallography. Does this mean that we no longer need those expensive and time-consuming methods? Well, for several reasons, this is not quite the case.
First and most importantly, predictions are not reality. They can help to simplify things or let us work into the right direction, but they cannot incorporate all the details of real biology. AlphaFold just takes a protein’s sequence of amino acids into account, but in reality, proteins are surrounded by water, small molecules and other proteins, and all of these affect the fold as well. Depending on the environment and on interaction with other molecules, some proteins even switch between multiple folds that are quite stable. Therefore, AlphaFold predictions are only a small part of the complete picture.
Another problem is posed by the so-called membrane proteins, a whole class of proteins which fix themselves at a membrane like, for example, the wall of a cell. Although AlphaFold does predict the individual parts across the membrane correctly, those are rarely aligned to each other correctly. This alignment problem occurs also with very huge proteins consisting of many smaller folded parts.
Last but not least, there are many proteins out there which were not predicted correctly by AlphaFold, so there’s still some room for improvement.
Important to mention is also the fact that AlphaFold was trained on the PDB, a database of experimentally solved protein structures. Without any new experimental data, prediction software like AlphaFold cannot be improved.
In summary, AlphaFold is good, but it is still not perfect.
In any case, the new predictions are useful not only to get a first glimpse of an unknown structure, but also to help solve structures by conventional methods. Remember that protein crystals are not always easy to grow? Well, this is sometimes due to regions which do not form a stable fold. AlphaFold2 can predict those regions and, thus, helps to design more successful experiments.
All in all, it is an incredible tool which does not only generate knowledge in a matter of hours instead of days but can also be used by everyone with no need for laboratory access or being an expert in structural biology. From here, we can only be curious what the next generation of structure prediction will be capable of, as AlphaFold2 already helped scientist around the globe to reveal the structural mysteries of various proteins.
