The instructions and files below will allow you to create your own model of the virus! All you need is some spare time and a 3D printer. In addition, those without access to a 3D printer can still use the STL files to request printing from external services and then follow the instructions on assembling the same way. We do hope that this model will make the virus more tangible and that the model will not only be printed as a private project, but also be used for outreach activities and in educational institutions.
These instructions refer to the updated SARS-CoV-2 3D model released in early 2022 that considers new scientific insights and improves on the original. You can find details about the changes (and our reasons for them) in another blog post soon.
Our design is based on the best scientific evidence available. Not only are the shapes of the various proteins as close to the measured molecular structures as possible, but their numbers as well as the overall size of the virion match experimental results on a scale of 1:1,000,000. Therefore, 1 mm on the model represents 1 nm (10 Å).
By the way, this would make the RNA that is inside the virus hull 10 meters long and 1 mm thick.
What you need to get started
For easier printing and assembly, the virus structure has been broken down into individual components:
For the model with rigid spikes:
https://www.thingiverse.com/thing:5326932/files
- Virion (one Top and one Bottom)
The two virion components are completely solid irregular hemispheroids. The body has been broken down into two separate parts in order to produce two flat surfaces, minimizing the need for supports and lowering the amount of excess material when printing. The outer surfaces contain the necessary recesses for the spike proteins to sit in. This model contains 26 such holes. The surface of the virion is textured to represent SARS-CoV-2's E and M proteins.


- 26 Spike Proteins
The spike protein stucture provides perhaps the most challenging aspect of the printing operation. Each spike consists of a complex crown-like surface and supporting stalk which connects it to the central virion. The individual spike STL files show the spike in different conformations and at varying angles.






For the model with springs:
https://www.thingiverse.com/thing:5666273/files
- Virion (one Top and one Bottom)
The two virion components are completely solid irregular hemispheroids. The body has been broken down into two separate parts in order to produce two flat surfaces, minimizing the need for supports and lowering the amount of excess material when printing. The outer surfaces contain the necessary recesses for the spike proteins to sit in. This model contains 26 such holes. The surface of the virion is textured to represent SARS-CoV-2's E and M proteins.


- 26 Spike Proteins (11 extended, 15 retracted)
The spike protein stucture provides perhaps the most challenging aspect of the printing operation. Each spike consists of a complex crown-like surface that is mounted on a spring.


- Springs
You can purchase a suitable spring at https://www.mcmaster.com/compression-spring-stock/od~1-8/id~0-085/. This spring is 20 inches or 508 mm long. You can cut it either in 26 19mm pieces or 28 18mm pieces with two spare ones. If you want to get your springs from some other source, make sure that their outer diameter is about the same as this one (0.125 inches or 3.2 mm). The springs have to fit into the holes in the model parts.
To date, the structures have been printed successfully on several Fused Deposition Modelling (FDM) printers (Rostok MAX v2 & Prusa I3 MK3 printers), and we anticipate the even higher quality structures will be feasible with alternative methods, such as stereolithography (watch this space). Each of the parts is available in STL format and is printable through any suitable slicer software. Personal discretion is advised when setting up the prints, as the exact details may differ depending on conditions and equipment. The procedure outlined below will serve as a good starting point. Let us know of your experience in the comments!
Printing of the component parts
The first step is to print the individual components. For the body parts this is very straight forward as the surface negates the need for supports. The body objects can be printed with the minimum infill for support, though infill of 10% is recommended for rigidity.
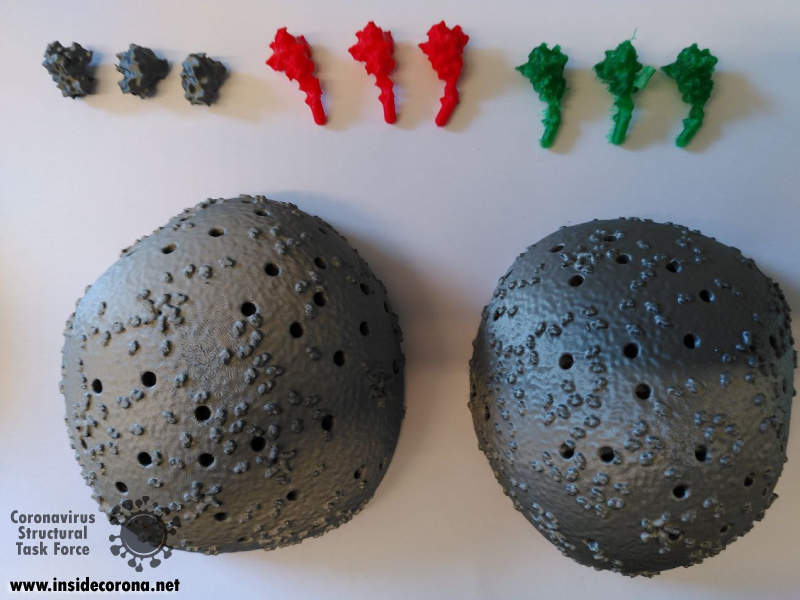
The spike proteins provide a more challenging print. The spike protein must be printed 26 times to complete the model. To represent the variety of conformations among the proteins in any given moment, we provide spikes in three different tilt angles in both the extended and the retracted state each. For a mixture of spike proteins that reflects a real-life virion reasonably well, we recommend this distribution:
For the model with rigid spikes:
- 3x 30° extended
- 4x 30° retracted
- 5x 40° extended
- 7x 40° retracted
- 3x 50° extended
- 4x 50° retracted
For the model with springs:
- 11x extended
- 15x retracted
The springs have to be about 3.2 mm in outer diameter and 18-19 mm in length. We recommend using stainless steel springs. To bend the springs, they are pulled onto several solid wires, which are bent into different spiral shapes. The wires with springs are then placed on a hot plate at 250 °C for 30 minutes. By using this method, different, random spring angles can be realized and the springs can be prevented from bending back into their original position.
We recommend printing the spike protein lying sideways as this results in stronger stalks. It is not too difficult to remove the supports of the stalks without breaking them.
We used FDM printing and ubiquitous poly-lactic acid (PLA), which made the post-processing easier.
A dual extruder printer would be ideal for spike printing as it would allow supports to be printed with water-soluble plastic, speeding up post-processing. In any case, printing individual or at least fewer spikes with greater spacing generally produces nicer objects which are easier to work with at the price of longer printing time.
Post-processing
Regardless of the approach taken for printing, some tidying will typically be needed to get the virion ready for assembly. Removing the supports can be done with a pair of pliers, while the smaller artifacts and issues will need brushing off or sanding. A dental pick can be quite useful.
For PLA, we found the best thing to clean and smooth the surfaces (after support removal) is ethyl acetate. While ethyl acetate is readily available in many chemical labs or at a pharmacy, acetone-free nail-polish remover offers a commercially accessible alternative. You should be using safety glasses and accurately fitting (!) gloves when handling ethyl acetate, ventilate the room well and, in case of skin contact, use a skin cream after washing your hands! It dissolves the plastic, breaking down the small extrusion artifacts on the surfaces and can be applied in many ways. We found it best to leave the parts in a sealed ethyl acetate vapor environment, e.g. by putting it in a smaller open vessel into a stainless-steel pot, which should be cleaned carefully afterwards. This technique results in even and clean results, though it will take up to a few days to fully smooth each object. The faster method is to simply submerge the small objects in ethyl acetate for 10-30 seconds, and then remove each object, leaving them to dry out on a surface. For the larger virion parts, the surface can be smoothed by rubbing it down with a cloth damped with ethyl acetate, which was also used to “weld” the two viral hull halves parts together. A small amount was dropped onto the flat surfaces on each section, before the two were pressed together until the plastic fused to become a single object. The seam was then smoothed down using the same process as before.
For acrylonitrile butadiene styrene (ABS), acetone may produce the same results.
Ready for Assembly!
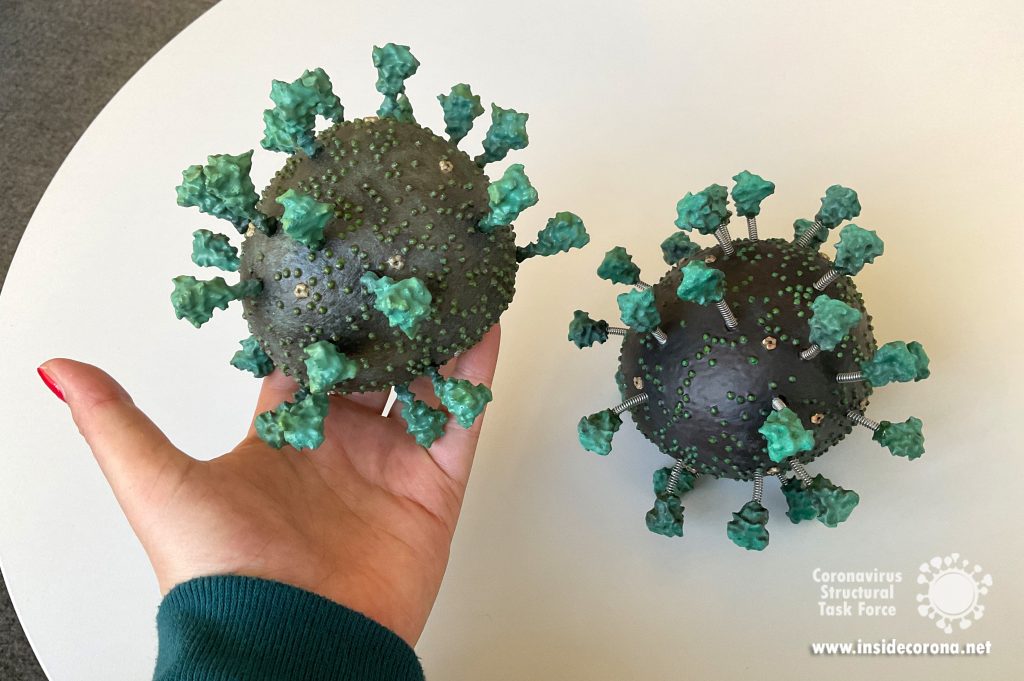
Finally, the 3D model can be assembled. For assembling, the springs are first fixed with superglue in the holes of the body. UV resin is then used for their final fixation. The UV resin also serves as a filling material to completely close the holes. The spikes are attached to the springs in the same way as the springs are attached to the body. If the intention is to paint the model, we recommend assembling the model before starting with the coloring. In this way, the UV resin used to fill the holes can also be painted and a more visually appealing result can be achieved.
We hope that our adventure in 3D printing the coronavirus inspires you to give it a try! The process we described was completed in a little over a week. The printing jobs were completed in just over two days, the cleaning and post-processing took another two days, while the painting was done over the course of a weekend. This article provides a description of our technique and should provide enough detail on how, with the outlined necessary tools, you can create a similar result. The files have been distributed through Thingiverse under a Creative Commons BY-NC license: You may remix, adapt, and build upon this work non-commercially and acknowledge the "Coronavirus Structural Task Force" as original author.
https://www.thingiverse.com/thing:5326932
As with every 3D printed model, there are many different ways this could be tackled and achieved, and we look forward to seeing the many creative ways explored by others in this endeavor. Please do share experiences and results with us, either through the comments Thingiverse or on Twitter (you can tag us @thornlab).

We have also designed a scale model of the human anti-body that binds to the spike protein. This is available alongside the virus model and can be attached to the spike protein as desired.
For a sense of perspective, we have also produced a model of the rhinovirus, which is one of the viruses that cause the common cold, at the same scale. It is available in STL format here: https://www.thingiverse.com/thing:4556845
Authors
We want to emphasize that the writing of this blog entry was a collaboration of a several people:
Dale Tronrud and Thomas Splettstößer worked together to create the STL files for the 3D model. Dale was the person to suggest it first (with Andrea Thorn picking up on the idea). Thomas then selected the experimental models and placed all the parts to form a realistic representation. Dale provided the knowledge about the limitations imposed by the nature of 3D printing and broke up Thomas’ model into printable parts that can be assembled without too much difficulty. He printed and assembled the first virion from this design. The updated model was printed at the facilities of the Physics department at the Universität Hamburg with generous support from PhysNET and Martin Stieben. Yunyun Gao and Philip Wehling refined the model, and Matthias Stäb painted the one shown in the pictures.
