Pharmaceutical drugs can be found by chance, but today, most so-called active pharmaceutical ingredients (APIs) are developed through a long, iterative process of designing and testing them.
How?
Targets and active ingredients
Most medicinal drugs are small molecules with up to 70 atoms, which bind in the body to larger molecules, or macromolecules. These so-called targets are typically proteins (long chains of amino acids), RNA, DNA (long chains of nucleotides) or carbohydrates (long chains of sugars). However, most targets are proteins.
Proteins are the workhorses of all living organisms: fungi, animals, plants, bacteria and even viruses (sic!) utilize them. They are the tools that allow us to digest what we eat, enable cell division, and form muscles and hair. One reason why our diet needs to contain proteins is that they are disassembled into amino acids from which new proteins can be made. Some amino acids we can’t make ourselves, so we HAVE to get them from our diet - these are the “essential” amino acids. To properly function, our body needs to be able to use a large variety of proteins - our genes encode at least 20,000 of them1!
An example: How Aspirin works
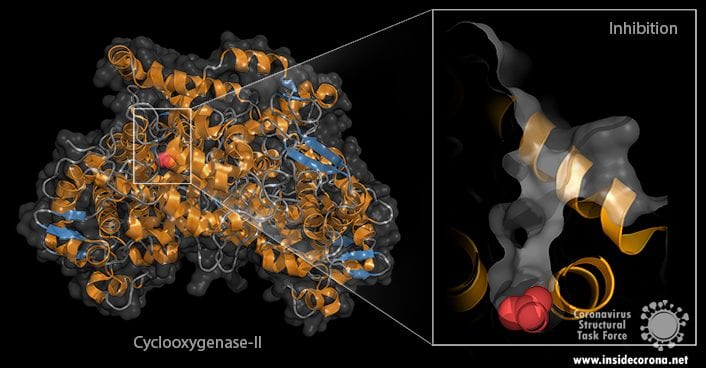
Our bodies require the proper function of proteins to live, but not all proteins are good for you: For chronic illnesses or metabolic disorders, diminishing the activity of certain proteins might be advantageous. A good example is cyclooxygenase-II or COX for short. This protein is formed when cells are injured or during inflammation and it catalyzes an important step in the production of pain mediators, called prostaglandines2. Without cyclooxygenase-II, no pain mediators can be produced.
Acetylsalicylic acid - also known as ASS or Aspirin – binds to cyclooxygenase-II and stops it from working (see image below). As a consequence, your body stops producing those pain mediators, and your pain is relieved*. In this case, cyclooxygenase-II is the target and ASS the active pharmaceutical ingredient.
Rational drug design
There are two major methods in rational drug design:
Indirect or ligand-based drug design utilizes molecules which are similar to known active pharmaceutical ingredients in their shape and charge. These molecules are then tested for binding and/or inhibition of the target, or, just as often, a resulting change in some biological parameter of interest, such as the killing of a virus!
Direct or structure-based drug design utilizes knowledge about the target. The potential API is chosen to bind to the target - for example, in the case of Cyclooxgenase-II, to fit in the channel (see image). In fragment based drug design, several smaller molecules are bound to sites in the target and with this knowledge, an active pharmaceutical ingredient is designed that combines their properties.
For both of these methods a pre-selection of potential molecules can be done by computer-aided drug design. However, for strutcure-based drug design, the macromolecular structure of the target must be known.
The Coronavirus Structural Task Force supports the search for a drug against COVID-19 by validating and, where possible, improving the macromolecular structures of potential coronavirus targets. We also offer drug designers information about different SARS-Cov-2 and SARS-CoV macromolecules. With this, we hope to do our part in the fight against COVID-19.
(blog header image: aspirin tablets by Ragesoss, Wikimedia Commons / license: CC 4.0)
* Unfortunately, instead of prostaglandines, the body then produces more leucotrienes, which can cause asthma attacks - hence, asthma patients should only take aspirin after consulting their GP.
- 1.Ponomarenko EA, Poverennaya EV, Ilgisonis EV, et al. The Size of the Human Proteome: The Width and Depth. International Journal of Analytical Chemistry. Published online 2016:1-6. doi:10.1155/2016/7436849
- 2.Ricciotti E, FitzGerald GA. Prostaglandins and Inflammation. Arterioscler Thromb Vasc Biol. Published online May 2011:986-1000. doi:10.1161/atvbaha.110.207449
