SARS-CoV-2: Not new, but different
The novel Coronavirus (2019‐nCoV) is classified as a large positive sense single stranded RNA-Virus from the family of betacoronaviruses. It shows high genetic similarity to SARS‐CoV and MERS‐CoV and is even closer related to the Bat-SARS-like corona virus, from which it most likely evolved. Even though it shows a lot of similarities to its ancestors, further insights in the infection mechanism and the structure of its proteins reveal significant differences.
But what is in it?
Like most RNA-viruses, the virus has a lipidic hull, with envelope and other proteins integrated in it. This viral shell is responsible for the interaction with host cells and the protection of the inner parts, most importantly: the viral RNA. This RNA acts as a direct template for the translation of two polyproteins named pp1a and pp1ab which encode the 16 non-structural proteins (nsps) of the replication‐transcription complex (RTC). Those 16 nsps, encoded by about two third of the genome (in terms of length), are cleaved from the polyprotein by the chymotrypsin‐like protease (3CLpro) (=Main protease) and one or two papain‐like proteases to generate the functional single proteins. As a result, the RTC synthesizes a variety of subgenomic RNAs (sgRNAs) in a discontinuous transcription, which serve as templates to produce subgenomic mRNA. Other open reading frames of the genome encode at least four structural proteins, that are necessary for the assembly of the virions, the hull and the infection of cells (called S-, M-,E- and N-protein for spike, membrane, envelope and nucleocapsid).
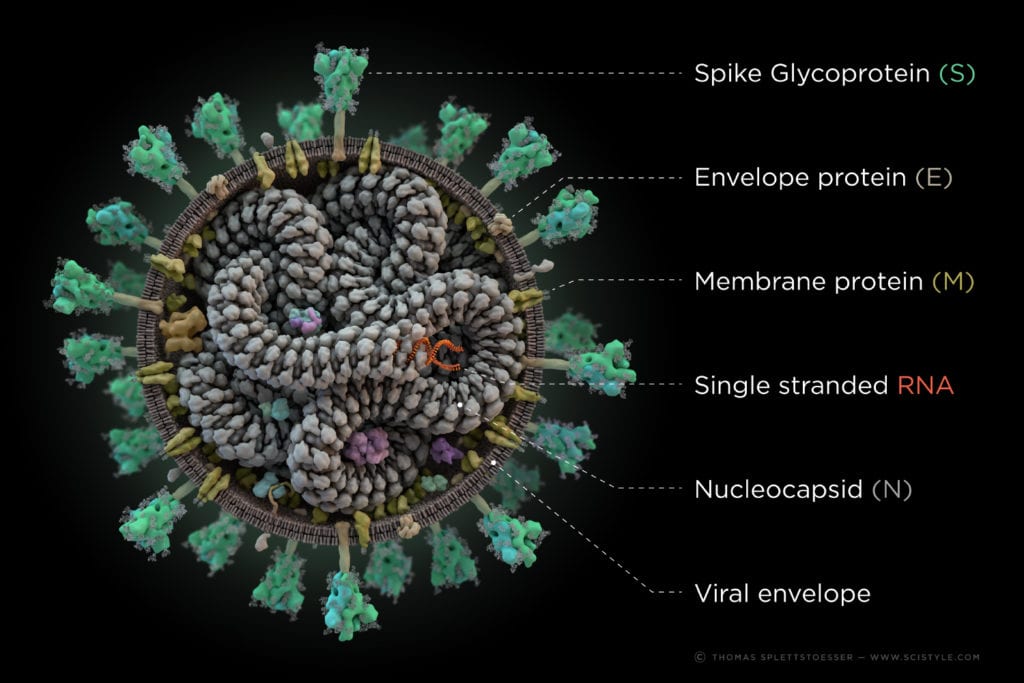
Making contact
The majority of infected cells are ACE2 (Angiotensin-converting enzyme 2)-bearing cells of the respiratory system. The viral mRNA is introduced through endocytosis via the spike glycoprotein of the Coronavirus. What does this mean? The S- or spike protein which forms the "corona" around the virus binds with its receptor-binding domain (RBD) to the receptor, which is located on the surface of the host cells. Afterwards, the virus can merge with the cell through a complicated mechanism, the so-called endocytosis. Once infected, these cells now act as a multiplicator for the virus which provokes a strong reaction of the immune system. Most common symptoms include cough, fever, fatigue, loss of taste, headache, diarrhoea, dyspnoea, and lymphopenia or pneumonia, even causing death of the patient in severe cases.
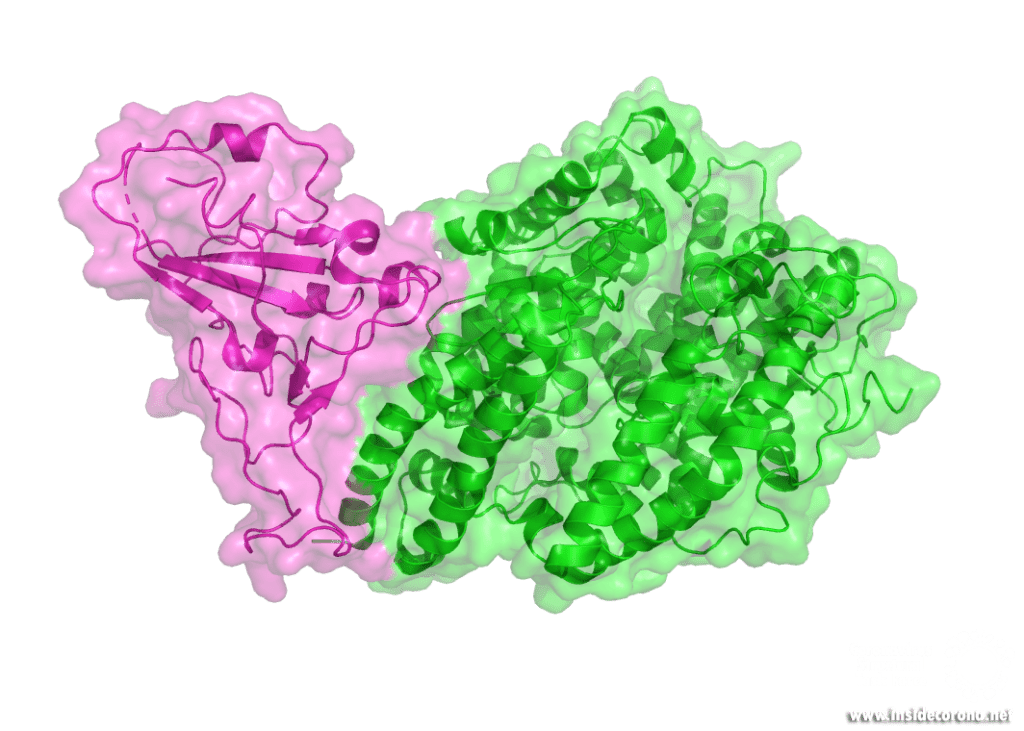
The structure of the virus, its infection mechanism and multiplication offer numerous possibilities for drug targeting, such as the inhibition of the main protease or the polymerases, the disturbance of the assembly of shell and entry proteins or the replication‐transcription complex and direct mRNA antiviral methods. However, none of them has been proven effective in clinical studies to this point.
Learn more:
- https://onlinelibrary.wiley.com/doi/pdf/10.1002/jmv.25681
- https://www.sciencedirect.com/science/article/pii/S0896841120300469
- https://www.nature.com/articles/pr198040
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1829448/
- https://onlinelibrary.wiley.com/doi/full/10.1002/jmv.25685
- https://en.wikipedia.org/wiki/Angiotensin-converting_enzyme_2
