“The coronavirus has led to a worldwide crisis for over a year. In a new study, nanoscientist Prof. Dr. Roland Wiesendanger illuminates the origins of the virus. His findings conclude there are a number of quality sources indicating a laboratory accident at the Wuhan Institute of Virology as the cause of the current pandemic.”
This is the beginning of an official press release from the University of Hamburg. Unfortunately, this study is not a study at all, but a rather confusing piece of internet research. And because we, the Coronavirus Structural Task Force, conduct research at the very same University of Hamburg, we would like to comment on the press release and this "study".

Here are the main arguments of the author, Roland Wiesendanger:
#1: Failure to identify the interim host proves that the disease is not of animal origin.
“In contrast to early coronavirus-based epidemics such as SARS and MERS, the scientific community has yet to identify the interim host that made the transmission of SARS-CoV-2 from bats to humans possible. Thus, there is no sound basis for a zoonotic theory as a possible explanation for the pandemic.” That an interim host of a zoonosis is not immediately identified is not unusual. The route of transmission of SARS-CoV was not elucidated until more than three years after the SARS pandemic [1]; for MERS, which was first described in 2012, it took two years [2] and to date some information is still missing [2–4]. Evidence indicates that interim hosts for SARS-CoV-2 may have been snakes, turtles, or pangolins [5–7]. Furthermore, transmission between humans and various animal hosts has been demonstrated several times (which, for example, unfortunately led to the culling of many mink)[8]. The lack of knowledge of an intermediate host does in no way disprove zoonosis as a cause.
#2: SARS-CoV-2 is so adapted to humans that it could not have arisen naturally
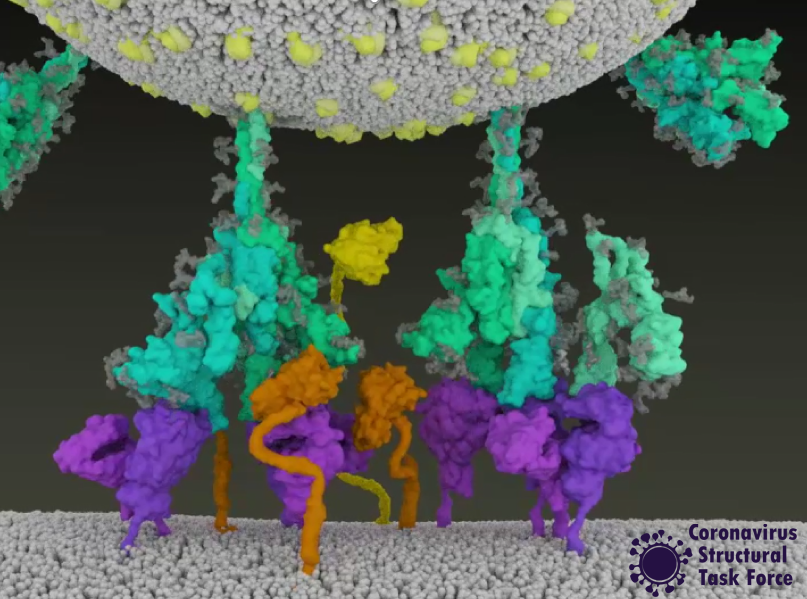
“The SARS-CoV-2 viruses are astonishingly effective at binding to human cell receptors and infecting human cells, thanks to its special cell receptor binding domains combined with a special (furin) cleavage site of the coronavirus spike protein. This is the first time a coronavirus has had both of these characteristics and indicates a nonnatural origin of the SARS-CoV-2 pathogen.”
The ability of the virus to bind to human cells with the spike protein is not evidence that this was artificially produced. Influenza, HIV, and Ebola are also all very good at binding to human cells [9] - the latter two have been shown to be animal-derived (zoonotic) pathogens. All of these viruses possess a furin cleavage site. Furins are enzymes found in all vertebrates that allow viruses to better target vertebrate cells. Thus, it is not unusual for SARS-CoV-2 to have evolved such a site through natural mutation and selection. In fact, it occurs naturally in many other coronaviruses as well [10].
The receptor binding domains S1 and S2 are highly variable because it is exactly domains that enable specific binding to host cells - the new mutations that are currently causing us so much trouble display variations right here. Mutations in this domain arise due to selection pressure in humans (or other hosts) and are also not indicative of a non-natural origin [11,12].
What also speaks against the virus being completely man-made, is that the sequence of the virus does not fit known methods for artificially producing genetic material [13,14]. Furthermore, designing a coronavirus would be considerably costlier than designing many other viruses because the genome is so large. This however, does not exclude the virus originating from a "gain-of-function" study.
Here it is perhaps worth noting that Professor Wiesendanger authored the study alone and is himself a non-specialist [15]. He has never published on Corona before, and therefore it is understandable that he is unfamiliar with both the details of genetic engineering and the technical terminology.
#3: Bats do not fly 2000km
“There were no bats for sale at the wet market in the center of Wuhan, which is the suspected hub of the outbreak. The Wuhan Institute of Virology, however, houses one of the largest collections of bat pathogens in the world, taken from distant caves in southern Chinese provinces. It is extremely unlikely that bats naturally made their way to Wuhan, from almost 2,000 km away, to then start a worldwide pandemic in the immediate vicinity of the Wuhan Institute of Virology.”
Considering that the thesis supported by WHO [16] is that there must have been an intermediate host, as is indeed also stated in the study and the press release (see 1.), the presence of bats near the first human vectors is not necessary. Many of the possible intermediate hosts were traded in the market in question. So far, there is no certainty that the pandemic originated there - research is still ongoing - but the absence of bats does not argue against zoonosis. The collection of bat viruses at the Center for Emerging Infectious Diseases in Wuhan does exist however. A researcher at this institute - Shi Zhengli - discovered that SARS originated from bats, and she conducted a systematic study of bat viruses from fecal samples in 2013, for example [17]. While the samples in question have been found in a cave 2000 kms from Wuhan, the bat in question (Rhinolophus affinis) does occur far and wide, including 250 km from Wuhan, in Hunan province. Bats have long been considered the largest source of different coronaviruses and thus the greatest risk for their transmission to humans [18].
#4: Research on viruses as bioweapons was conducted in Wuhan
“One research group at the Wuhan Institute of Virology had been researching the genetic manipulation of coronaviruses for many years with the goal of making these more infectious, more dangerous, and more fatal. This has been demonstrated by numerous publications.”
The publications cited in the study, for example this one [19], are indeed concerned with the recombination of bat coronaviruses with spikes that can bind to human cells. These were used to trace how the 2002/3 SARS pandemic originated - not to make the virus more dangerous. Such research has taken place elsewhere - with many strict safety measures and safeguards - for example in North Carolina [20].
#5: The virological institute in Wuhan was not secure.
“Safety measures were documented as being insufficient at the Wuhan Institute of Virology prior to the outbreak of the coronavirus pandemic.”
The laboratory in Wuhan is a Biosafety Level 4 laboratory [21], the highest level - there are only a handful of such laboratories in the world, of which two are in China. Such labs have strict access controls, you have to be able to hermetically seal them off, and they are under negative pressure to prevent pathogens from escaping; access is only through an airlock; all wastewater is chemically and thermally treated; a full protective suit has to be worn, and when leaving, the whole body has to be cleaned with soap. Of course, no laboratory is perfect, but safety is a paramount issue in such laboratories [21]. Nature has written a special report on the lab in Wuhan that illustrates this. In the text, Prof. Wiesendanger argues with a serious article from the Washington Post that points to episodes of malpractice in the laboratory in Wuhan, but whose sources remain undisclosed. Another source, a Youtube video, supposedly proves the improper disposal of laboratory equipment. However, while garbage can be clearly seen, typical laboratory waste such as pipette tips, consumables and gloves, is missing. Furthermore, the Chinese text of the video does not mention the waste in any way, it is about whether and how one could get into the building. A second source [22] shows a bat allegedly being sampled without protective equipment. Parts of the video, which is about a new coronavirus in pigs, show the non-BSL-4 area of the lab. The new coronavirus in pigs is said to come from the bat species shown. Nothing in this video suggests biosecurity problems in the lab.
6. Authorities covered up a laboratory accident in October
“There are numerous direct indications that the SARS-CoV-2 pathogen is of laboratory origin and point to a young researcher at the Wuhan Institute of Virology as being the first person to be infected. In addition, there are indications that the SARS-CoV-2 pathogen emerged from the Wuhan Institute of Virology into the city of Wuhan and beyond. There are also indications that the Chinese authorities conducted an examination of the institute in the first half of October 2019.”
The "indications of an official examination in the first half of October" are based on an analysis of mobile phone location data that an external firm is said to have produced for the Pentagon to show disruption of laboratory operations and road closures. However, the report provides no concrete evidence and was therefore deemed insufficient by the intelligence community. Especially since some of the allegations could be directly refuted [23]. Little can be found on the Internet about the young scientist, Yan Ling Huan, apart from a Twitter account, videos, and a rebuttal from the lab. The hypothesis that she was the first to be infected can therefore neither be proven nor disproven.
All in all, one cannot rule out the lab as the point of origin - research is being done on coronaviruses there - but the sources cited in this "study" are not evidence of that. Circulation of the virus before December cannot be ruled out either, but the publication does not present any robust evidence.
Additional notes
The main problem with this study is that it appears to have been written by the author alone and was not peer reviewed. This is particularly regrettable since the author emphasizes the importance of peer review on page 3. Apparently, discussion and especially media attention are very desirable, but peer review is not, which is why the "publication" was done on the Research Gate platform, where you can simply upload a PDF.

Other problems with the "study" are the poor readability and deficiencies in the evidence - this document is not only confusing, but also does not comply with good scientific practice. Sources include not only private communications and Twitter posts, but also content from the Alt-Right movement, such as an unrefereed study [24,25] from Steve Bannon's entourage, articles from the Epoch Times [26] or Summit News [22]. The document is also full of contradictions – for example, the first patient is said to have fallen ill on the first of October 2019, but elsewhere it says that the pandemic is due to a laboratory accident between October 6 and 11, 2019. The use of colored markers in the text also does not necessarily contribute to readability and is uncommon in publications. It is not clear why such a highly decorated and well-known scientist as Prof. Wiesendanger considered such writing publishable or even a study; and, unlike many of his colleagues, he does not do research on the coronavirus.
Conclusion
What comes across as an invitation to debate is a rather disorderly and angled piece of internet research that does not correspond with good scientific practice. Many people who are against China, or who are simply looking for someone to blame for Corona, feel vindicated and the University of Hamburg backs this up.
It is good and right that professors at German universities can publish and research whatever they want ("Forschungsfreiheit"). But the fact that this article is published in close consultation with the president [15] but without peer review and in the name of the university does not cast a good light on the University of Hamburg, which is, after all, our very own scientific home. Our statements here are about the shortcomings of this press release as well as the paper, and are not intended as criticism of Prof. Wiesendanger personally; we deeply regret this press release. As scientists, we should educate, critically question - and allow ourselves to be questioned. For the last twelve months, we have been informing colleagues and the general public about ongoing corona research, taken care not to accept funding from potential influencers (e.g. the pharmaceutical industry), and carefully reviewed every blog post, no matter how small. Every day we respond to inquiries from people who are confused, afraid of corona or of the measures being put in place. We teach, educate, we answer questions.
And that is what we will continue to do.
I would like to thank Dr. Florian Platzmann, Dr. Sam Horrell, Dr. Yunyun Gao, Pairoh Seeliger, Lea von Soosten, Katharina Hoffmann, Joshua Ezika and Sabrina Stäb for their help writing this article. Their expertise in lab safety, public health care, mandarin, molecular biology and their comprehensive internet / literature research made this opinion piece possible.
- [1]Li W, Wong S-K, Li F, et al. Animal Origins of the Severe Acute Respiratory Syndrome Coronavirus: Insight from ACE2-S-Protein Interactions. JVI 2006: 4211–4219. doi:10.1128/jvi.80.9.4211-4219.2006
- [2]Han H-J, Yu H, Yu X-J. Evidence for zoonotic origins of Middle East respiratory syndrome coronavirus. Journal of General Virology 2016: 274–280. doi:10.1099/jgv.0.000342
- [3]Middle East respiratory syndrome coronavirus (MERS-CoV). . Im Internet: https://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov); Stand: 19.02.2021
- [4]Chen F, Cao S, Xin J, et al. Ten years after SARS: where was the virus from? J Thorac Dis 2013; 5 Suppl 2: S163–S167. doi:10.3978/j.issn.2072-1439.2013.06.09
- [5]Ji W, Wang W, Zhao X, et al. Cross‐species transmission of the newly identified coronavirus 2019‐nCoV. J Med Virol 2020: 433–440. doi:10.1002/jmv.25682
- [6]Liu Z, Xiao X, Wei X, et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS‐CoV‐2. J Med Virol 2020: 595–601. doi:10.1002/jmv.25726
- [7]Zhao J, Cui W, Tian B. The Potential Intermediate Hosts for SARS-CoV-2. Front Microbiol 2020. doi:10.3389/fmicb.2020.580137
- [8]Mahdy MAA, Younis W, Ewaida Z. An Overview of SARS-CoV-2 and Animal Infection. Front Vet Sci 2020. doi:10.3389/fvets.2020.596391
- [9]Becker GL, Lu Y, Hardes K, et al. Highly Potent Inhibitors of Proprotein Convertase Furin as Potential Drugs for Treatment of Infectious Diseases. Journal of Biological Chemistry 2012: 21992–22003. doi:10.1074/jbc.m111.332643
- [10]Wu Y, Zhao S. Furin cleavage sites naturally occur in coronaviruses. Stem Cell Research 2021: 102115. doi:10.1016/j.scr.2020.102115
- [11]Lau S-Y, Wang P, Mok BW-Y, et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerging Microbes & Infections 2020: 837–842. doi:10.1080/22221751.2020.1756700
- [12]Liu Z, Zheng H, Lin H, et al. Identification of Common Deletions in the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus 2. J Virol 2020. doi:10.1128/jvi.00790-20
- [13]Almazán F, Sola I, Zuñiga S, et al. Coronavirus reverse genetic systems: Infectious clones and replicons. Virus Research 2014: 262–270. doi:10.1016/j.virusres.2014.05.026
- [14]Andersen KG, Rambaut A, Lipkin WI, et al. The proximal origin of SARS-CoV-2. Nat Med 2020: 450–452. doi:10.1038/s41591-020-0820-9
- [15]Ursprung des Coronavirus: Uni mit fragwürdiger Theorie. . Im Internet: https://www.zdf.de/nachrichten/politik/corona-labortheorie-universitaet-hamburg-100.html; Stand: 19.02.2021
- [16]WHO-convened Global Study of the Origins of SARS-CoV-2. . Im Internet: https://www.who.int/publications/m/item/who-convened-global-study-of-the-origins-of-sars-cov-2; Stand: 19.02.2021
- [17]Ge X-Y, Li J-L, Yang X-L, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013: 535–538. doi:10.1038/nature12711
- [18]Anthony SJ, Johnson CK, Greig DJ, et al. Global patterns in coronavirus diversity. Virus Evolution 2017. doi:10.1093/ve/vex012
- [19]Ren W, Qu X, Li W, et al. Difference in Receptor Usage between Severe Acute Respiratory Syndrome (SARS) Coronavirus and SARS-Like Coronavirus of Bat Origin. JVI 2007: 1899–1907. doi:10.1128/jvi.01085-07
- [20]Menachery VD, Yount BL Jr, Debbink K, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 2015: 1508–1513. doi:10.1038/nm.3985
- [21]Cyranoski D. Inside the Chinese lab poised to study world’s most dangerous pathogens. Nature 2017: 399–400. doi:10.1038/nature.2017.21487
- [22]Video From 2018 Shows Chinese Scientists Working On Coronavirus In Wuhan Lab. . Im Internet: https://summit.news/2020/04/30/video-from-2018-shows-chinese-scientists-working-on-coronavirus-in-wuhan-lab; Stand: 19.02.2021
- [23]Report says data suggests shutdown at China lab, but experts skeptical. NBC News. Im Internet: https://www.nbcnews.com/politics/national-security/report-says-cellphone-data-suggests-october-shutdown-wuhan-lab-experts-n1202716; Stand: 19.02.2021
- [24]Coronavirus: Wie Steve Bannon die Labor-Theorie verbreitet. . Im Internet: https://www.zdf.de/nachrichten/panorama/coronavirus-wuhan-labor-studie-yan-bannon-100.html; Stand: 19.02.2021
- [25]Yan L-M, Kang S, Guan J, et al. Unusual Features of the SARS-CoV-2 Genome Suggesting Sophisticated Laboratory Modification Rather Than Natural Evolution and Delineation of Its Probable Synthetic Route. 2020. doi:10.5281/ZENODO.4028830
- [26]Exclusive: China Had COVID-Like Patients Months Before Official Timeline. The Epoch Times (Singapore) 2020. Im Internet: https://epochtimes.today/exclusive-china-had-covid-like-patients-months-before-official-timeline/; Stand: 19.02.2021

All Roland Weisendanger presented is his scientific judgment, which is at the same level as those who judged it is not from lab. It goes without saying, open a thought should never be forbidden, but rule out a possibility must be careful.
https://de.wikipedia.org/wiki/Institut_f%C3%BCr_Virologie_Wuhan
Der Ursprung des Virus ist doch so einfach das noch nicht einmal darüber diskutiert werden muß? Ein Viruslabor in U
Wuhan mit der Aufgabe tierische Viren für den Mesnchen scharf zu schalten - und hier wird tatsächlich noch darüber diskutiert?
Dear both, thanks for this informative and reliable, i.e. scientific, discussion of Roland's article.