Our goal was to create a 3D printable model of SARS-CoV-2 that is as close to the actual virus as possible, but there are a lot of confusing and contradictory descriptions of the coronavirus SARS-CoV-2 in the literature. Combing through them in order to establish a complete understanding and a clear image of the virus that causes COVID-19 is a tremendous task and as new scientific observations come to light, our conceptions are being challenged, and need for adjustments arises.
At the start of the pandemic, when there were few measurements of the novel virus, its appearance was mainly inferred from knowledge about other coronaviruses, especially the closely related SARS-CoV-1 which caused the previous SARS pandemic [1]. The Coronavirus Structural Task Force created a printable model based on those images, but since then a lot of imaging data has been collected on the new virus, itself, and those images indicate that SARS-CoV-2 is sufficiently different. We have combed the literature and used what we learned to design an updated model.
The Shape of the Virus
Because viruses are so small, it is very difficult to directly observe them even with modern imaging techniques. (For more details, see here.) In the olden days, viruses were studied by coating samples with metal and then taking an electron microscopic image (see figure) so that the surface can be clearly seen. This is, by the way, the origin of the name “coronavirus”: The viral particles looked a bit like little suns surrounded by a corona of rays of light.
The most powerful ways to study the shape of a virus do not just investigate a single particle, but average the images of hundreds of them together resulting in information that does not describe a single virion but instead their average. In reality, and particularly for coronaviruses, every individual virus particle looks different, some being larger, and some smaller. They are only perfectly round in the absence of any exterior forces or perturbing internal structure. It does not take much to deform the “wobbly" thin double membrane hull of SARS-CoV-2. For example, it has been suggested that coronavirus particles are created in a close-to-perfect spherical shape and exposure to a slightly acidic environment causes virions to become a bit deformed, a change that might be important for infectiousness, and it has been postulated that the conformation of the M proteins affects the membrane curvature and, hence, the shape of the virus [2]. Our model is therefore not exactly round but more shaped like a potato. We do not claim that this is “the shape” of the virus, but simply one of many possible shapes.
A second difference between the old and new data is that SARS-CoV-2 appears to be smaller than the other viruses studied before this pandemic. To reflect the new data we have reduced the diameter of our model by 12%, from 100 mm to 88 mm. (Our model is scaled so 1 mm corresponds to 1 nm.) As with virual shape, individual viruses have a variety of diameters and 88 mm is simply one of many sizes which are consistent with the population. Due to the adjustments to the virus body its hull surface was reduced by roughly 23%. Research suggests that M proteins are distributed on the surface of a virion with a roughly constant density so we reduced their number by a matching 23%. The data for the E protein, however, indicates a roughly constant number in each virion so we have left their number at twenty. At this scale, the RNA contained in the virion would be about 10 meters long and one mm thick. One of the models for the virion body that we supply has a hole in the bottom that, both, allows the model to sit on a table and be used as a paper weight, and allows a 10 m long piece of twine to be hidden inside. The twine can be extracted during a demonstration to emphasize the surprising length of the SARS-CoV-2 RNA molecule.
Reconsidering the Spikes
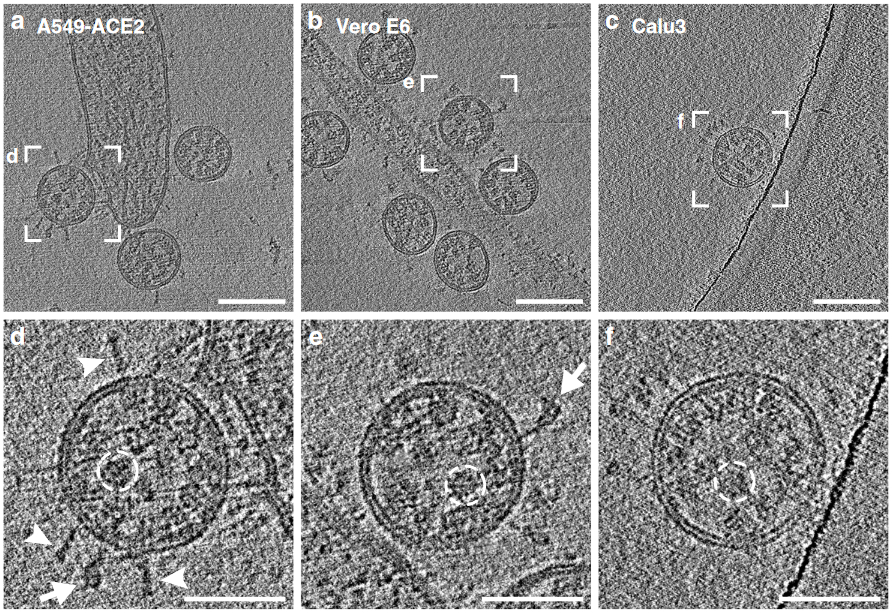
As was found for the size of the virion, the experimental data derived from direct study of SARS-CoV-2 indicates a different number of spikes per particle than seen in previous studies, and the new number is far smaller. Current literature suggests the average number of spikes on the SARS-CoV-2 viral surface ranges from 26 [3] to 48 [4] . Our first model of SARS-CoV-2, based on images of other coronaviruses, had around 100 spikes but in our new model we have reduced their number to only 26.
While it is possible that SARS-CoV-2 does, indeed, have a different number of spikes than other coronaviruses, it is also possible that this substantial difference might well stem from changes in experimental approaches and circumstances. The microscopes and imaging techniques of today are much more powerful than those of even ten years ago.
In the past, despite the inconceivably small size of a single virion, coronavirus particles were too large to for all of its parts to be in focus in an image. Since one could only “see” the spikes in a limited region of a virus some rather elaborate methods were employed to estimate the total number of spikes. One of these is the Tammes problem, a mathematical puzzle that aims to distribute the maximum number of nonoverlapping, equal-sized circles on the surface of a sphere. Estimates for the minimal distance between two spike proteins on the viral surface are readily available even from 2D imaging, and following the assumption that the entire surface of the virion is covered with spikes packed as closely as possible the Tammes method suggests the presence of around 50 spikes per virus particle.

However, many images of SARS-CoV-2 show gaps between spikes, and it is quite likely that the density of spikes is lower. New techniques applied to SARS-CoV-2 can produce a sharp image of an entire, individual particle allowing the spikes to simply be counted. These studies show an average of 26 spikes per virion and this is probably the most reasonable estimate available until further experimental evidence emerges. This is just an average; the exact number of spikes on a particular virion varies around that value. The same publication also states that spikes are able to rotate freely on the viral surface and are not standing entirely upright, but are instead inclined by 40° on average [3], again varying a lot from spike to spike. Our updated version of the SARS-CoV-2 model incorporates these new findings. It has 26 spikes whose stalks are bent at angles of 30°, 40° and 50°. To improve the model’s flexibility and sturdiness, we also produced a version where the spike tops are connected to the body via springs. However, enthusiasts should remember that the spike proteins (as well as membrane and envelope proteins) are also “swimming” in the bilipidic membrane that serves as the outer hull, which we could not represent in our model.

Now, where’s the Model?
We made the updated files for the 3D-printable virus model available on thingiverse.
In this blog article you can find detailed construction guidance for the new model.
And here's a 3D preview of the new model on Sketchfab:
References
[1] "Coronavirus never before seen in humans is the cause of SARS". United Nations World Health Organization. 2003-04-16. Archived from the original on 2004-08-12. Retrieved 2022-02-17.
[2] Neuman, B., Kiss, G., Kunding, A. H. et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 174, 1 (2011), 11–22. https://dx.doi.org/10.1016%2Fj.jsb.2010.11.021.
[3] Yao, H., Song, Y., Chen, Y. et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 183, 3 (2020), 730-738. https://doi.org/10.1016/j.cell.2020.09.018.
[4] Klein, S., Cortese, M., Winter, S.L. et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun 11, 5885 (2020). https://doi.org/10.1038/s41467-020-19619-7.
